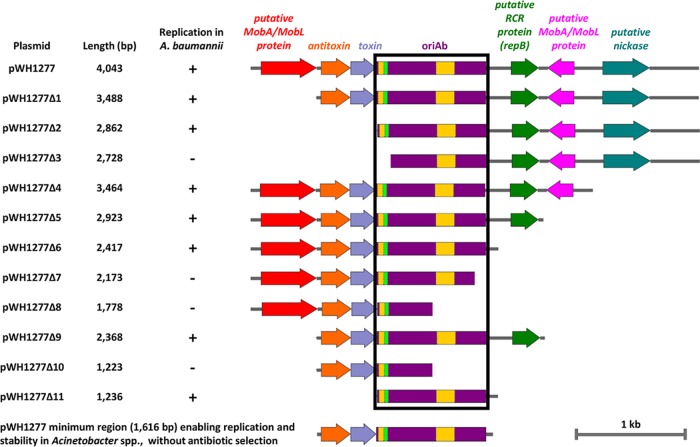
FIG 1.
Deletion analysis of pWH1277 to determine the minimal region required for autonomous plasmid replication in Acinetobacter spp. Deletion fragments of pWH1277 (24) were generated by PCR amplification with primers listed in Table 1 and blunt cloned at the SmaI site of pBS. The resulting pWH1277 deletion derivatives were introduced in A. baumannii ATCC 19606T to assess replication. The minimal self-replicating region of pWH1277 (oriAb) is boxed in black. Relevant coding regions are indicated with colors: red and pink, two putative genes coding MobA/MobL-like proteins, presumably involved in plasmid mobilization; orange, the PaaA2-like antitoxin component of the TA system involved in plasmid stability; cobalt, the ParE2-like toxin component of the TA system involved in plasmid stability; violet, the oriAb origin of replication for Acinetobacter spp., including a palindromic sequence (fluorescent green) and two A+T-rich regions (yellow); green, the gene coding for a putative RepB-like protein, presumably involved in rolling-circle replication (RCR); cyan, the gene encoding a putative nickase, likely involved in single-strand DNA nicking for rolling-circle replication. All genes are reported in scale over the total length of the vector. Images were obtained by the Snapgene software (GSL Biotech).