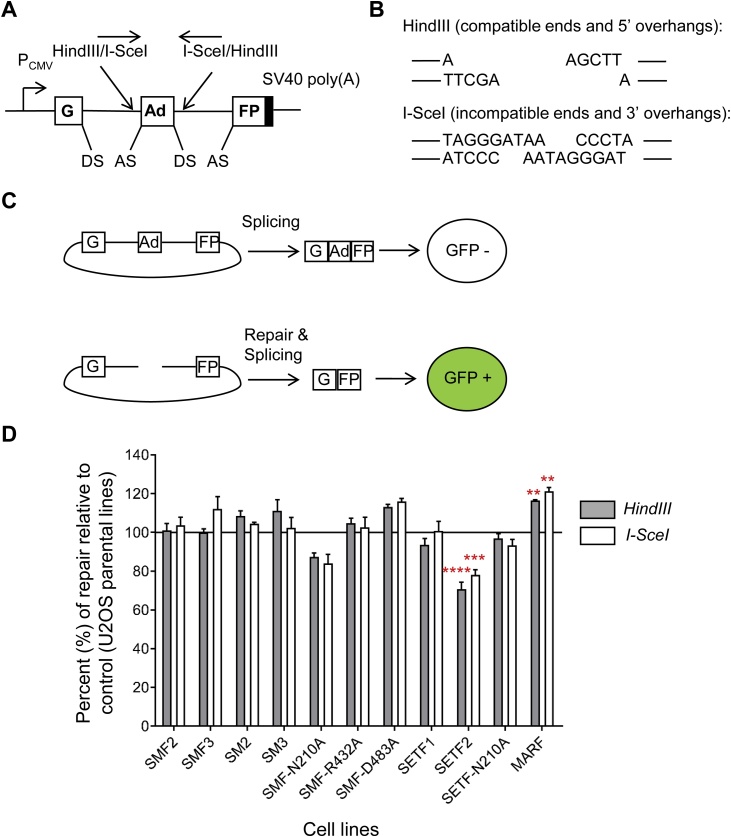
Fig. 5.
The SET and MAR domains have an opposite effect on DNA repair.
A, The reporter plasmid, pEGFP-Pem1-Ad2, is composed of a GFP cassette flanked by a PCMV promoter and a SV40 poly(A) sequence. The eGFP coding sequence is interrupted by a 2.4 kb intron containing an adenovirus exon (Ad). The Ad exon is flanked by HindIII and I-SceI restriction sites. The donor (DS) and acceptor (AS) splicing sites are shown. B, HindIII and I-SceI restriction sites are respectively composed of a palindromic 6-bp and a non-palindromic 18-bp sequence. Digestion of the reporter by HindIII or I-SceI generates respectively compatible and incompatible ends. C, The presence of the Ad exon in the eGFP ORF inactivates the eGFP activity thus making the cell eGFP negative. Removal of the Ad exon by HindIII or I-SceI followed by a successful intracellular repair will restore eGFP expression, which is then quantified by flow cytometry. The assay was adapted from ref. [33]. D, The frequency of end joining in the indicated cell lines compared to the parental U2OS and U2OS-F cell lines was determined by flow cytometry. Average ± S.E.M. of 3 biological replicates. Statistical test: one-way ANOVA Dunnett’s multiple comparisons test, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001.