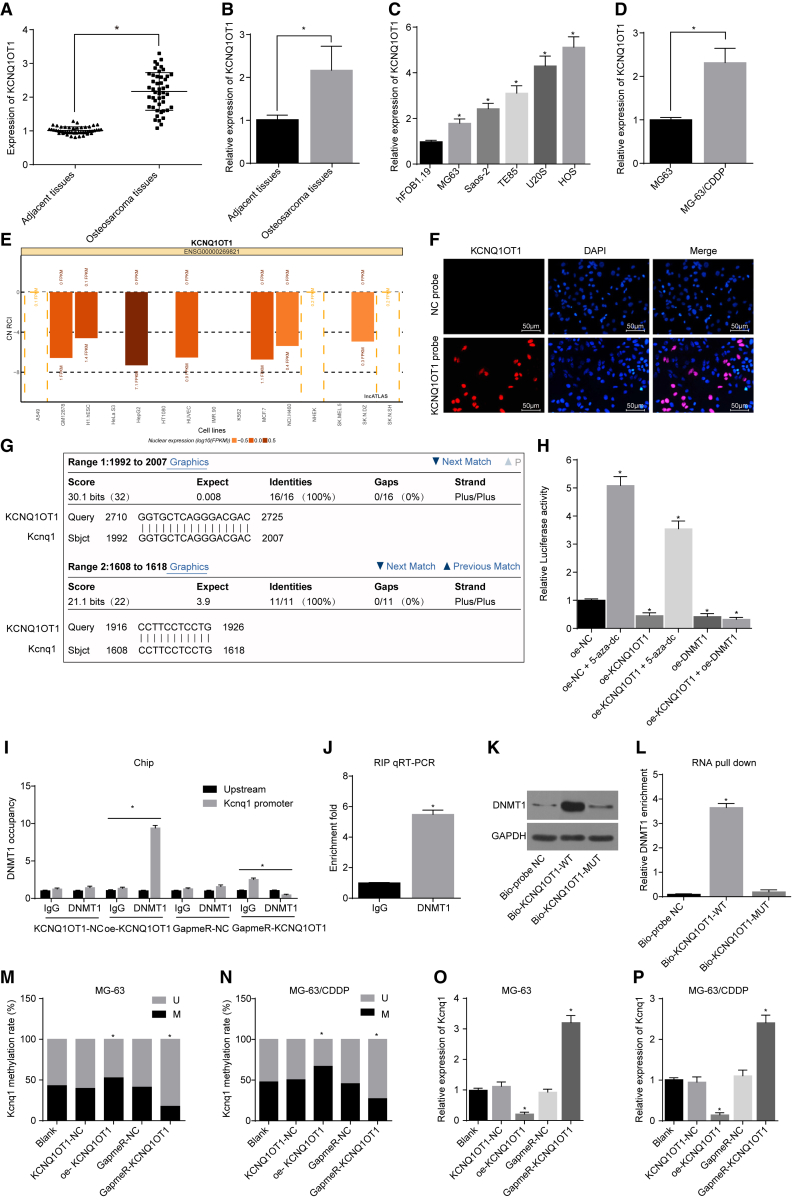
Figure 5.
The Correlation between Kcnq1 Expression and KCNQ1OT1 in Osteosarcoma Cells
(A and B) KCNQ1OT1 expression in osteosarcoma cells and normal cells. (C) KCNQ1OT1 expression in each osteosarcoma cell line. (D) KCNQ1OT1 expression in the MG-63 cell line and MG-63/CDDP cell line. (E) The subcellular localization of KCNQ1OT1 predicted on the lncATLAS website. (F) The subcellular localization of KCNQ1OT1 (200×). (G) Similarities between KCNQ1OT1 and Kcnq1 gene promoter, compared using BLAST. (H) Luciferase activity in each group. (I) The enrichment of DNA methyltransferase DNMT1 in the Kcnq1 promoter region. (J) The effect of KCNQ1OT1 on the enrichment of DNA methyltransferase DNMT1. (K and L) The effect of KCNQ1OT1 on pulling down DNMT1 protein. MG-63 and MG-63/CDDP cells were treated with oe-KCNQ1OT1 or GapmeR-KCNQ1OT1, with KCNQ1OT1-NC and GapmeR-NC as the controls. (M and N) The level of Kcnq1 methylation in each group. (O and P) The mRNA expression of Kcnq1 in each group, determined by qRT-PCR. *p < 0.05 versus the normal group, the hFOB1.19 cell line, the MG-63 cell line, the NC group, the blank group, the IgG group, or the Bio-probe NC group. The measurement data were expressed as mean ± SD. Comparison between two groups was analyzed by independent t test, and comparisons among multiple groups were processed with one-way ANOVA. The experiment was repeated 3 times. ChIP, chromatin immunoprecipitation; FISH, fluorescence in situ hybridization; BSP, bisulfite sequencing PCR; RIP, RNA immunoprecipitation; 5-Aza-dC, 5-Aza-2 deoxycytidine; NC, negative control; IC50, inhibitory concentration 50%; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; WT, wild-type; KCNQ1OT1, KCNQ1 opposite strand/antisense transcript 1; DNMT1, DNA methyltransferase 1.