Abstract
The surface of proteins is heterogeneous with sophisticated but precise hydrophobic and hydrophilic patches, which is essential for their diverse biological functions. To emulate such distinct surface patterns on macromolecules, we used rigid spherical synthetic dendrimers (polyphenylene dendrimers) to provide controlled amphiphilic surface patches with molecular precision. We identified an optimal spatial arrangement of these patches on certain dendrimers that enabled their interaction with human adenovirus 5 (Ad5). Patchy dendrimers bound to the surface of Ad5 formed a synthetic polymer corona that greatly altered various host interactions of Ad5 as well as in vivo distribution. The dendrimer corona (1) improved the ability of Ad5-derived gene transfer vectors to transduce cells deficient for the primary Ad5 cell membrane receptor and (2) modulated the binding of Ad5 to blood coagulation factor X, one of the most critical virus–host interactions in the bloodstream. It significantly enhanced the transduction efficiency of Ad5 while also protecting it from neutralization by natural antibodies and the complement system in human whole blood. Ad5 with a synthetic dendrimer corona revealed profoundly altered in vivo distribution, improved transduction of heart, and dampened vector sequestration by liver and spleen. We propose the design of bioactive polymers that bind protein surfaces solely based on their amphiphilic surface patches and protect against a naturally occurring protein corona, which is highly attractive to improve Ad5-based in vivo gene therapy applications.
Keywords: polyphenylene dendrimer, amphiphilic surface patches, adenovirus, dendrimer−virus complexes, dendrimer corona, gene delivery, gene therapy
Only recently, it has been discovered that the protein corona largely determines the fate of nanoparticles in the bloodstream in vivo.1−5 Unlike highly specific ligand–receptor recognition, protein corona formation on nanoparticle surfaces mainly depends on multiple weak molecular binding events.4 Protein surfaces are not homogeneous but provide defined amphiphilic patterns. Although the role of such amphiphilic surface patches in cell biology is still barely understood, it has been found essential for their cellular distribution and various interactions with, for example, other proteins, membranes, and carbohydrates.6 In consequence, molecular recognition concepts have been proposed such as the corona phase model based on folded heteropolymers constrained at a single-walled nanotube surface,7 which indicate that slight polarity differences could induce large differences in molecular biorecognition. The importance of amphiphilic patches has been previously demonstrated in a synthetic model, where gold nanoparticles have been prepared with alternating hydrophobic and hydrophilic segments; these nanoparticles penetrated cellular membranes more efficiently than particles with the same functional groups homogeneously distributed on the surface.8 Therefore, we hypothesized that by engineering specific amphiphilic patches, one could control biomolecule recognition and even engineer a protein-like corona by synthetic macromolecules.
In nature, viruses are among the most intelligent and efficient nanotransporters. Their enormous efficiency not only depends on their highly specific receptor binding, but they can also utilize blood plasma proteins as shielding corona to enhance their stability and evade the immune system.9 Specific binding of blood plasma proteins is predetermined by the surface of the virus capsid. For instance, blood coagulation factor X (FX) binds to the surface of adenovirus type 5 (Ad5) forming a protein corona that shields the viruses from attack by natural antibodies and complement,9 which also significantly enhances their liver transduction efficiency.10,11 In clinical trials, Ad5 is the most frequently used gene transfer vector to date12 by providing high transduction efficiency in both dividing and nondividing cells and accommodating large transgene cassettes.13 However, the clinical applicability of Ad5-based vectors is severely limited by this native protein corona formation9,10,14 since the absorbed proteins dictate whether the vector can transduce certain cells in vivo and frequently lead to mistargeting and acute toxicity.15 For example, the FX in the protein corona triggers highly efficient sequestration of Ad vector particles in the liver,10,11 which is a major limitation in Ad5-based gene therapy. In addition, while Ad5 in vitro employs the coxsackie–adenovirus cell surface receptor (CAR) and α(v)β3/5 integrins to infect cells, the primary receptor CAR is often not available on target tissue in vivo,16 and tumor cells are often refractory to transduction by Ad5.17 Therefore, it would be a significant advancement for Ad-based gene therapy if one could engineer an artificial protein-like corona that replaces the natural protein corona, which should shield the vector from neutralizing antibodies and ultimately engineer virus tropism for therapeutic applications.
Our goal was to mimic the complex amphiphilic patterns like the surface topology of proteins by shape-persistent polyphenylene dendrimers (PPDs) to ultimately control biorecognition of Ad5. PPDs were prepared, providing alternating negatively charged sulfonic acid and hydrophobic n-propyl groups (e.g., amphiphilic groups) on their surfaces,18,19 and we have identified PPD3 (see Scheme 1) to provide an optimal amphiphilic surface texture for Ad5 vector binding. PPD3 forms a protein-like corona on Ad5 and alters Ad5 biorecognition and pathways for gene transduction. We could demonstrate enhanced vector transduction into low CAR cells, protecting Ad5 from FX binding and plasma neutralization, and alter the in vivo tissue distribution of Ad5. Our results are of great relevance to improve in vivo gene therapy by synthetic polymers, providing precise amphiphilic surface patterns. We further envision that our concept could be utilized to create nanoparticle therapeutics with tailored amphiphilic surface patterns that serve as artificial protein corona to control the in vivo distribution of nanoparticles, which represents a holey grail in nanomedicine.
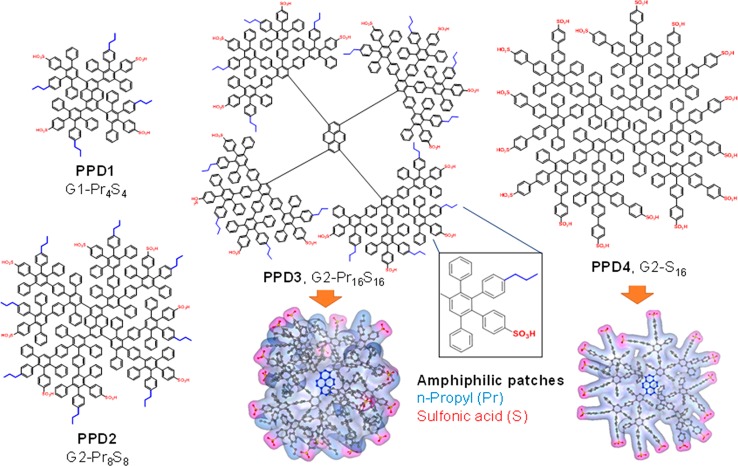
Scheme 1. Synthesized PPDs with Amphiphilic n-Propyl and Sulfonic Acid Surface Patches.
Variation of the sizes (1st generation G1-Pr4S4, “PPD1” vs 2nd generation, e.g., G2-Pr8S8 or PPD2), negative charges as well as surface patches. Visualization of the 3D structure of patchy PPD3 and PPD4.
Results and Discussion
Dendrimers with Precise Amphiphilic Surface Patches
In cell biochemistry, sulfonic acid groups are only present in certain carbohydrates such as heparin. Inspired by heparin sulfate–glycosaminoglycans, which are observed in blood and undergo multiple protein–protein interactions, we have synthesized PPDs with surface-exposed sulfonic acid surface groups. In contrast to carboxylic acids, sulfonic acid groups are negatively charged independent of the pH (within the biologically relevant range). As lipophilic residues, propyl chains were selected due to their performance in a previous in vitro screening that analyzed the influence of various nonpolar substituents (i.e., phenyl, propyl, isopropyl, butyl, and hexyl chains) on the cell uptake and toxicity of PPDs.20 1,3,6,8-Tetraethynylpyrene served as the dendrimer core so that the blue emission of pyrene could be used as a fluorescence probe to enable monitoring of the macromolecules via fluorescence microscopy. PPDs with different but precisely established numbers and orientations of the sulfonic acid/n-propyl surface groups were synthesized by reacting the core with functionalized cyclopentadienone building blocks in Diels–Alder cycloadditions. Detailed characterization of each sample is also given in the Supporting Information Section 2 (Scheme 1).21 We then considered the impact of the size (first vs second generation), surface-charge density (PPD2 vs PPD3), and significance of the n-propyl groups (PPD3 vs PPD4) on the interactions between the “patched” surface dendrimers and Ad5.
In the following sections, we describe the formation of PPD3/Ad5 complexes and their transduction pathways by a combination of biological and physicochemical methods. The key concern is to alter the surface topology of Ad5 by dendrimer binding to ultimately control and improve critical interactions with plasma proteins and cells.
PPD3/Ad5 Complexes with Improved Transduction Efficiency
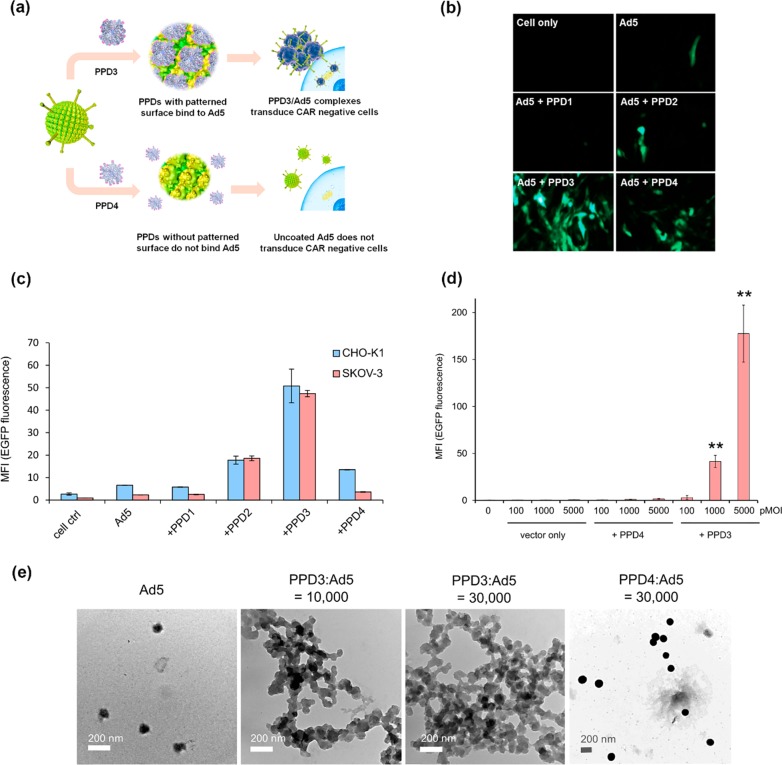
PPD1 to PPD4 were mixed with Ad5 at a desired ratio in PBS buffer, and the gene transduction capabilities of Ad5 in the uncoated and complexed form were tested. Replication-defective Ad5-based vectors harboring a heterologous expression cassette for the enhanced green fluorescent protein (EGFP) were used as model system to study transduction of different cell lines (see SI for details, Ad5 mentioned in the experimental and results sections always refers to this vector). Vector particles and PPDs were incubated in PBS buffer at a molar ratio of 1:10 000, and the complexes were subsequently used for transduction. After 24 h post transduction, EGFP was quantified by flow cytometry. CHO-K1 (a subclone of Hamster Chinese ovary noncancer cell line) and SKOV-3 (human ovarian cancer cell line) cells were selected, which are difficult to transduce by Ad5 due to low CAR receptor expression. The corresponding flow cytometry data are depicted in Figure 1c. From PPD1 to PPD3, with increasing numbers of alternating sulfonic acid/propyl groups, the transduction enhancement is increased accordingly with PPD3/Ad5 complexes exhibiting the highest transduction efficiencies, about 20-fold higher than Ad5 control alone. The presence of amphiphilic surface patches was crucial for bioactivity since PPD4 with similar size and the same number of acid groups as PPD3, but lacking the n-propyl chains, revealed almost no increase of transduction efficiency for SKOV-3 cells. The trends seen for different dendrimers were also directly visualized by fluorescence microscopy (Figure 1b and Figure S3). With increasing virus concentrations (particle multiplicity of infection = physical particles per cell, pMOI) from 1000 pMOI to 5000 pMOI, the transduction enhancement with PPD3 became even more prominent (>30-fold, Figure 1d). Clearly, biorecognition of Ad5 could be altered after PPD3 binding to the virus surface, and both the ratio of the amphiphilic groups (sulfonic acid and n-propyl) and their surface density on PPDs are essential for their interactions with Ad5.
Figure 1.
PPD3 and Ad5 formed complexes with increased transduction efficiency on CHO and SKOV-3 cells. (a) Illustration of PPD binding to Ad5 and transduction. (b) Fluorescence microscopy images of EGFP expression in CAR-negative CHO-K1 cells. PPD3 significantly enhanced Ad5 uptake and EGFP expression in comparison to other PPDs. (c) Flow cytometric quantification of Ad5-mediated EGFP expression in CAR-negative CHO-K1 and SKOV-3 cells after incubation with PPD1, PPD2, PPD3, and PPD4; MFI stands for mean fluorescence intensity as absolute values. Transduction was performed with 100 pMOI (particle multiplicity of infection). (d) Flow cytometric quantification of Ad5-mediated EGFP expression in SKOV-3 cell with different pMOI of Ad5 (n = 3). (e) TEM images (scale 200 nm) of Ad5 before and after incubation with PPD3 and PPD4. Complex formation between dendrimer and Ad5 only occurred upon incubation with PPD3. ∗∗ represents p-value ≤ 0.01.
This significant binding of PPD3 to Ad5 was therefore studied further by light scattering and electron microscopy (Figure 1e and Table S1). By increasing the ratio of PPD3 to Ad5, the particle sizes of formed complexes increased from 108 ± 11 nm (Ad5) up to about 729 ± 87 nm for PPD3/Ad5 (1000:1 ratio) (Table S1) but maintained low polydispersity index (PDI) below 0.2. In contrast, PPD4 with the same number of sulfonic acid groups as PPD3, but no n-propyl groups, did not form any complexes with Ad5 (Table S1) even at very high concentrations of the dendrimer (10 000:1). The PPD3/Ad5 complexes could be visualized by transmission electron microscopy (TEM) (Figure 1e), while, here again, no complexation of PPD4 with Ad5 was found. Obviously, PPD3 molecules formed a protein-corona-like structure on Ad5, and the result is in agreement with the transduction data and supported the assumption that defined patches of amphiphilic groups (sulfonic acid and n-propyl) are critical for promoting interactions with Ad5. The interaction between PPD3 and Ad5 was also detected by biolayer interferometry (BLI) (Figure S10, Figure S11, and Table S2). In the BLI experiment, Ad5 particles with high number of binding sites interact with a flat sensor surface densely coated with PPD3–dendrimer binding motifs. According to this method, the calculated KD (equilibrium dissociation constant) is as low as 1.27 pM. Most likely, this very strong binding is a result from multivalent interactions of PPD3 and Ad5, and it does not reflect the binding event of single PPD3 molecules with Ad5. However, we clearly show that strong interactions occur between this dendrimer and Ad5.
Enhanced Ad5 Transduction of Cells with Low CAR Expression
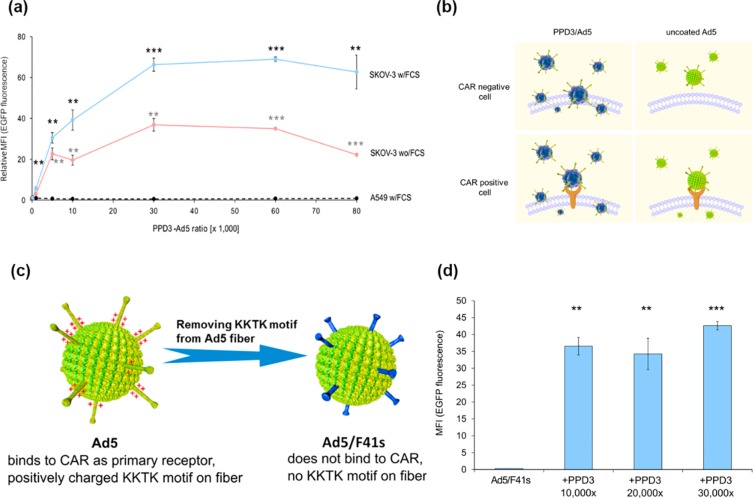
Coxsackie–adenovirus receptor (CAR) is the main receptor at the cellular membrane that recognizes and interacts with Ad5.22 Consistently, cells that do not express CAR (e.g., most tumor cells) can hardly be transduced by Ad5. By increasing the ratio of PPD3 to Ad5, we observed a saturable increase in the efficiency for Ad5 transduction of SKOV-3 cells with low CAR expression (Figure 2a). In contrast, transduction of A549 cells with high CAR levels was not influenced by increasing concentrations of PPD3 (Figure 2a). Similar observations were obtained with other high and low CAR cell lines (Figure S5). These results revealed that the formation of a PPD3 corona around Ad5 particles could promote an alternative, yet unknown pathway for Ad5 uptake that significantly improves Ad5 transduction into low CAR cells (Figure 2b).
Figure 2.
Transduction by PPD3/Ad5 complexes was independent of the primary Ad5 receptor CAR and the positively charged KKTK motif in the fiber capsomere. (a) In the presence of FCS, the transduction of SKOV-3 cells by PPD3/Ad5 complexes was increased (blue line) compared to the absence of FCS (red line) over a broad range of different PPD3/Ad5 ratios. Transduction of the human nonsmall cell lung carcinoma line A549 was not increased by PPD3/Ad5 complexes (dashed line). Transduction was performed with 1000 pMOI for all PPD3/Ad5 ratios. The relative MFI value calculated from transduction by Ad5 only is shown. (b) Illustration of the difference in cell uptake with PPD3/Ad5 and uncoated Ad5. PPD3/Ad5 uptake into CAR negative cells independent from CAR receptor, whereas uncoated Ad5 could not. For CAR positive cells, PPD3/Ad5 might still have utilized the CAR-binding pathway, thus resulting in the same transduction efficiency as uncoated Ad5. (c) Depiction of native Ad5 with positively charged KKTK motif in the fiber, binding CAR as primary receptor and recombinant Ad5/F41s without the positively charged KKTK motif in the fiber that does not bind CAR. (d) Transduction assay with Ad5/F41s and PPD3. Despite a lack of CAR binding, the KKTK motif PPD3/Ad5/F41s complexes revealed significantly enhanced transduction efficiency, which indicated the independence of both CAR and KKTK. ∗∗ represents p-value ≤ 0.01, ∗∗∗ represents p-value ≤ 0.001.
Enhanced Transduction in Serum
For in vivo applications, the impact of serum on the transduction efficacy of the PPD3/Ad5 complexes is crucial. As demonstrated in Figure 2a, the presence of fetal calf serum (FCS) did not reduce transduction efficiency of PPD3/Ad5 complexes; instead, we even observed a significantly increased Ad5 transduction efficiency. Increased transduction efficacy has been reported for DNA-containing lipoplexes23 due to a protein corona coating of the lipoplexes, which induced different cellular uptake pathways. In addition, it has also been shown that distinct corona proteins can enhance cellular binding and uptake.24,25 Therefore, to study the interaction between blood proteins and PPD3, the surface of a literature known model system, polystyrene nanoparticles (ø = 100 nm) were coated with PPD3, and the protein corona composition was compared to the uncoated nanoparticles. Via SDS-PAGE and proteomics (Figures S12–S14), we could demonstrate that the protein corona significantly changes after the nanoparticles were coated with PPD3. After incubation with blood serum, a significantly higher amount of vitronectin was bound the PPD3-coated nanoparticles in comparison to the uncoated ones. Vitronectin is one example of a corona protein that has been reported to mediate cellular interactions with cancer cells.26 Further studies are required to analyze the interaction with serum proteins in greater detail. In these studies, we could clearly demonstrate that serum proteins stabilized the PPD3/Ad5 complexes in blood serum, which is attractive within in vivo applications.
Transduction Independent of Primary Ad5 Receptor CAR and Positively Charged KKTK Motif in Fiber Capsomere
Since PPD3 exhibits a net negative charge, we initially speculated that the interaction of Ad5 and PPD3 could be of electrostatic nature involving positively charged amino acids on Ad5. The most prominent positively charged motif at the Ad5 surface is the KKTK motif located at the fiber shaft.27 Therefore, an Ad5 mutant Ad5/F41s lacking the KKTK and being unable to bind to CAR was investigated (Figure 2c). The structure of Ad5/F41s is almost identical to Ad5 with only a deletion of the KKTK motif on the fibers. PPD3/Ad5/F41s complexes demonstrated very high uptake into SKOV-3 cells, as indicated by high EGFP expression depicted in Figure 2d. This result, again, highlighted that compared to Ad5 alone, PPD3 enhanced vector transduction when the CAR pathway was not available. Also, PPD3 still interacted with Ad5/F41s efficiently despite the absence of the positively charged KKTK. Therefore, binding between PPD3 and Ad5 most likely occurred at a site distinct from the fiber and did not depend solely on electrostatic interactions between the negatively charged sulfonic acid groups of PPD3 and the positively charged KKTK sequence on Ad5. Instead, the main driving force for the dendrimer interaction with Ad5 could be the distinct alternating patches of hydrophilic negative charges and hydrophobic propyl groups of PPD3.
Dendrimer-Coated Ad5 Inhibits Transduction Pathway Mediated by Blood Coagulation Factor X (FX)
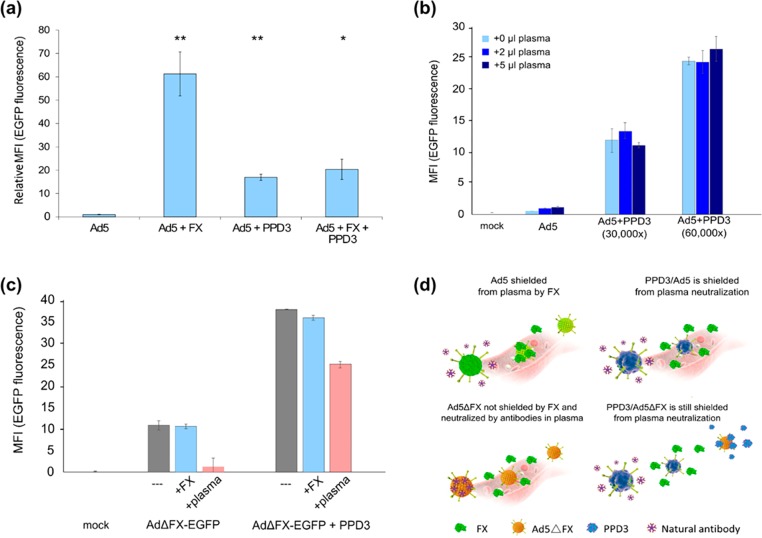
Factor X is a Vitamin K-dependent zymogen of the coagulation cascade. It is a serine endopeptidase, which is synthesized in the liver. Trimers of the Ad5 hexon protein bind with nanomolar affinity to FX, forming 240 FX binding sites per virion.10 Following intravenous (i.v.) administration of Ad5, viral particles predominantly accumulate in and transduce the liver in rodent and nonhuman primate models.28−30 This is a major limitation for Ad5-mediated gene therapy in humans. In vitro, FX functions as “bridge receptor” that facilitates attachment of Ad5 to cells. The γ-carboxyglutamic acid domain of FX binds Ad5, and the serine protease domain of FX binds cell-surface heparan sulfate proteoglycan (HSPG).10 Therefore, in agreement with previously published work,10 addition of FX to cell culture media could significantly enhance the transduction efficiency of Ad5 (Figure 3a). Surprisingly, after mixing of FX with PPD3/Ad5 complexes, no transduction enhancement was observed (Figure 3a), which suggested that PPD3 either occupied a similar binding site on Ad5 as FX or that it was bound to a site that could affect FX binding. In both cases, FX would be unable to enhance transduction of PPD3/Ad5 complexes. The results were corroborated by using superphysiological levels of FX, which are also unable to improve Ad5 transduction (data not shown). There is a competition between the PPD3–Ad5 and FX–Ad5 binding, which indicates that the FX and PPD3 sites are overlapping. FX is known to interact with the hexon capsomer; thus, it is very likely that PPD3 also binds to the hexon proteins. Since FX leads to liver transduction of Ad5 in vivo, using the PPD3 corona that replaces the FX corona holds promise for reducing liver toxicity and also to redirect Ad5-based gene delivery systems to other organs.
Figure 3.
PPD3/Ad5 complexes exhibit increased transduction efficiency in human plasma and PPD3 protected FX-binding ablated capsids from neutralization by the IgM/complement pathway. (a) PPD3/Ad5 complexes were formed and SKOV-3 cells transduced (1000 pMOI) in the presence or absence of FX at its physiological concentration (8 μg/mL). While FX was expectedly able to significantly increase transduction of SKOV-3 cells, the transduction by PPD3/Ad5 complexes was not enhanced by FX, suggesting that PPD3/Ad5 complexes may not be able to bind FX. (b) PPD3/Ad5 complexes exhibited increased transduction of largely refractory SKOV-3 cells in the presence of human plasma of an Ad-naïve donor (1000 pMOI). (c) FX is known to shield Ad5 particles from attack by natural IgM antibodies and complement. A FX-binding ablated Ad5 vector, a preferable tool for gene transfer and oncolysis due to its significantly reduced hepatotropism, did not show enhanced transduction on SKOV-3 cells (1000 pMOI) but was expectedly neutralized after addition of 5 μL of plasma from an Ad-naïve donor. PPD3/Ad complexes at a molar excess of 30 000-times were largely resistant to neutralization, indicating that PPD3 can replace FX as a protective shield against IgM/complement. (d) Comparison of uncoated Ad5, PPD3/Ad5, Ad5ΔFX, and PPD3/Ad5ΔFX for their FX binding and subsequent neutralizing antibody binding. ∗ represents p-value ≤ 0.05, ∗∗ represents p-value ≤ 0.01.
PPD3 Corona Protects Ad5 from Plasma Neutralization
To solve the challenge of FX-mediated sequestration of Ad vectors by hepatocytes, a number of strategies have been proposed including genetic modifications of the virus capsid to ablate FX-binding.31 However, Xu et al. have previously reported that elimination of FX binding enabled Ad neutralization by natural antibodies and complement. In this case, the vector was quickly neutralized due to the lack of the shielding by FX bound to the virus capsid.9 To analyze if PPD3 could shield the viruses from neutralizing natural antibodies and complement, we incubated the PPD3/Ad5 complex with the plasma from an Ad naïve human donor prior to transduction. The efficiencies, analyzed on SKOV-3 cells, did not decrease after plasma incubation (Figure 3b). In addition, we formed complexes between Ad5 vectors ablated for FX-binding (Ad5ΔFX) and PPD3 to demonstrate that PPD3 can prevent plasma neutralization even without FX binding. These complexes were incubated with human plasma. In the absence of PPD3, the vectors were completely neutralized by plasma (Figure 3c), while in the presence of PPD3, infectivity was preserved to a very large degree: PPD3 not only binds to FX-binding ablated vectors but also protects them from natural antibodies and complement (Figure 3d), which is essential for in vivo applications.
PPD3 Corona Does Not Affect Wild Type Ad5 (wt Ad5) Activity and Ad5 Replication
The above experiments utilized a replication-defective adenoviral vector leaving it unclear whether PPDs would hamper the ability of the virus to replicate. Any negative impact on virus replication would disqualify PPD3/Ad5 complexes for oncolytic cancer treatment. Therefore, we analyzed the potential of PPD3/wt Ad5 complexes to lyse tumor cell lines. Since wt Ad5 lyses tumor cells, the cytotoxicity assay was used to evaluate the activity of wt Ad5 (Figure S6). The formation of complexes did, however, not inhibit virus replication, but increased lysis of SKOV-3 cells compared to wt Ad5 alone again made the PPD3/Ad5 complexes suitable tools for oncolytic virotherapy.
Decreased Liver Transduction and Improved Heart Transduction in Vivo
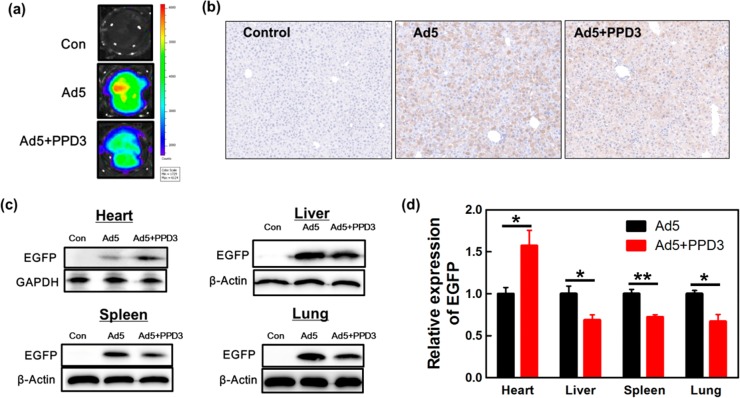
PPD3/Ad5 complexes were injected into male C57BL/6J mice at the age of 9 weeks, and transduction was analyzed in different organs 14 days post injection. We speculated that PPD3/Ad5 complexes would exhibit reduced liver transduction compared to naked Ad5. Therefore, the EGFP expression level in liver was initially assessed by fluorescence imaging of the whole liver tissue (Figure 4a) to demonstrate Ad5-mediated transduction. In fact, EGFP expression in liver tissue was reduced by 40% as supported by immunohistochemical analysis of EGFP in hepatocytes (Figure 4b). Similar results were also seen by quantification of EGFP DNA in liver using real-time quantitative PCR (Figure S8). Noteworthy, the liver was found undamaged in both cases (Figure S9). In addition, EGFP expression was quantified in liver, heart, spleen, and lung by Western blot analysis (Figure 4c,d), which demonstrated decreased transduction of liver and spleen, and lung by about 40% compared to naked Ad5. Very surprisingly and importantly, PPD3/Ad5 complexes showed increased transduction of heart tissue by more than 40%.
Figure 4.
Decreased liver and improved heart transduction by PPD3/Ad5 complexes in vivo. (a) Fluorescence imaging of the whole mice liver. (b) Immunohistochemical analysis of liver tissue (10× magnification). The cell nucleus was stained by hematoxylin indicated in blue; EGFP was stained by antibodies and DAB (3,3′-diaminobenzidine) as depicted by yellow color. (c) Western blot of EGFP expression in heart, liver, spleen, and lung. (d) Quantification of relative EGFP expression in different organs based on Western blot. All data for PPD3/Ad5 are normalized to the corresponding EGFP expression with naked Ad5 (n = 3). ∗ represents p-value ≤ 0.05, ∗∗ represents p-value ≤ 0.01.
Therapeutic Potential of Ad5 with Patchy Dendrimer Corona
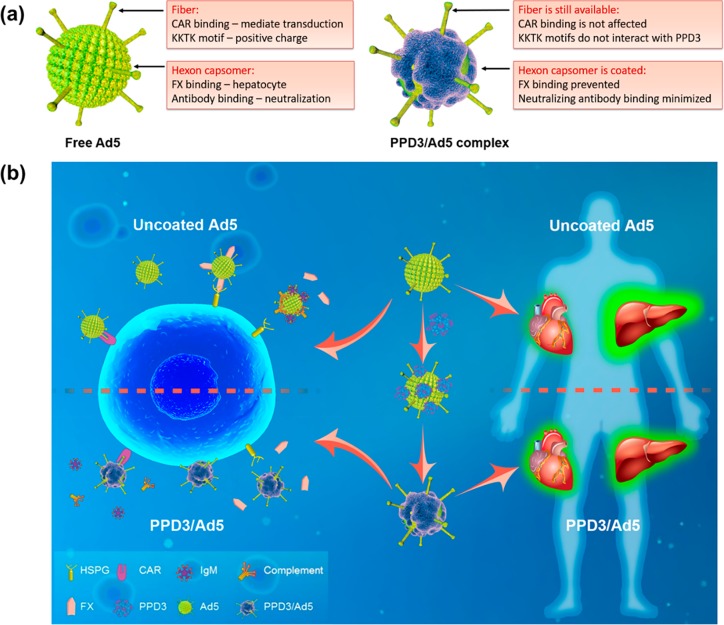
PPDs with a defined surface patterning of amphiphilic groups was able to specifically bind to the Ad5 surface, altering the existing surface topology of Ad5 and imparting a synthetic dendrimer corona providing different molecular recognition features (Figure 5a). In this way, the interaction of Ad5 with (a) its cellular CAR receptor, (b) plasma proteins, and (c) the coagulation factor X changes drastically. After PPD3 coating, the transduction mechanism of the PPD3 coated Ad5 becomes independent of naturally existing pathways such as CAR and FX, and it significantly prevents the immediate neutralization of the vector by the innate defense system composed of natural antibodies and complement. Firm evidence is provided that upon PPD3 binding, the conventional cellular uptake of Ad5 via CAR is changed, thus opening a different, still unknown pathway. This furnishes three significant advantages providing potential therapeutic opportunities. First, the complexes can traffic into CAR negative cells, which is not possible for Ad5 and Ad5/FX. This is highly attractive as many cancer or stem cells do not express CAR, and therefore, these cells cannot not be transduced by Ad5. Therefore, PPD3 holds great promise to widen the therapeutic options of Ad5 as depicted in Figure 5b.
Figure 5.
Schematic overview of the features of Ad5 with PPD3 corona. (a) Comparison of uncoated Ad5 and PPD3/Ad5 complex. CAR binding site and KKTK motif present on the fiber of naked Ad5, while FX and natural antibodies bind to the hexon. PPD3 forms a protein-mimicking corona on the virus capsid to prevent FX binding and vector neutralization. (b) Summary and visualization of PPD3 binding with Ad5, highlighting the impact of the PPD3 corona on cell Ad5 uptake processes and in vivo distribution (the figure is designed by the graphic designer Weihang Zhao and permission to use is granted).
Second, the interaction of Ad5 with serum proteins is altered. Intravenous delivery of adenovirus vectors requires that Ad5 is not inactivated in the bloodstream, and serum neutralizing activity is well documented for Ad5. In the case of Ad5 with PPD3 corona, serum proteins even boost gene transduction, which represents an essential feature for in vivo applications.
Third, PPD3 binding has a strong impact on Ad5 interactions with FX as both PPD3 and FX seem to compete for the same site at the surface of Ad5, and FX does not affect the transduction efficiency of preformed PPD3 coated Ad5.
Fourth, intravenous (i.v.) administration of Ad5 predominantly leads to accumulation of viral particles in the liver, and transduction occurs mainly in the liver. Therefore, several studies have been performed that document the role of blood coagulation FX in mediating liver gene transfer in vivo and the mechanism underlying Ad5 hepatocyte transduction10,14,15,32,33 by FX binding to the hexon protein of Ad5 at high affinity (∼2 nM), thus mediating hepatocyte transduction in mice and rats.10 In this way, gene therapy to other organs has been greatly limited, which can be addressed and improved by Ad5 with PPD3 corona (Figure 5b). Our in vivo studies proved that the PPD3 corona significantly reduced the Ad5-mediated transduction indicated by EGFP expression in the liver by about 40% but increase the transduction in the heart by more than 40% in comparison to naked Ad5. There is, thus, evidence that the PPD3 corona imparts a different biorecognition profile for Ad5, which alters the Ad5 tropism in vivo.
Conclusions
Our results demonstrate that rationally synthesized amphiphilic surface patterns of three-dimensional macromolecules play a critical role for their biological recognition and interaction with Ad5. The opportunity to remodel the virus surface by precise molecular and supramolecular design provides control over virus–host interactions.
Redirecting Ad5 tropism has been achieved by genetic engineering of Ad5. However, PPDs with defined amphiphilic patches provide a versatile strategy to modify the Ad5 surface and to reengineer Ad5 biodistribution. On the basis of this, one could further advance Ad5-based gene therapy toward higher tissue/organ specificity. Although the reason why the PPD3 complexes target the heart is still unknown, it is envisioned that by synthesizing PPDs with different tissue targeting moieties, tissue distribution of PPD/Ad5 complexes will be controlled by rational design. Therefore, we shall attach further tissue specific targeting groups to PPD structures to redirect Ad5 transduction to other organs. This will ultimately regulate the trafficking and cell uptake of viruses in vivo, one of the holey grails in gene therapy. Moreover, the concept of a bioactive protein-mimicking polymer corona could allow designing improved nanoparticle therapeutics to control their in vivo distribution, which is still one of the major challenges in nanomedicine.
Methods
Complex Formation of Ad5 and PPD3
Complex formation of Ad5 and PPD3 was performed in 50 μL of phosphate buffered saline (PBS). This volume contained 5 × 109 Ad5 virus particles (VP) that resulted in a concentration of 108 Ad5 particles per μL (the concentration of Ad5 was quantified by measuring the absorbance at 260 nm and converted to number of particles with the equation: concentration (VP/mL) = OD260 × 1.1 × 1012). PPD3 was added in defined ratios (if not otherwise mentioned, ratio of 1:10 000 was used in the experiments). The volume needed of the PPD 3 stock solution (c = 1 mg/mL, solved in DMSO) was calculated according to the following formula (N, number of particles; r, ratio between PPD3 and Ad5; M, molecular weight of PPD3; V, volume; NA, Avogadro constant; c, concentration):
This is a calculation example for a PPD3/Ad5 ratio of 1:10 000:
In this sample with a PPD3/Ad5 ratio of 1:10 000, 0.8 μL of the PPD3 stock solution was added to Ad5 particles and filled up with PBS to a volume of 50 μL. Before usage of PPD3, the stock solution was incubated in the sonicator for ∼30 min to dissolve assembled molecules because after some storage time, PPD3 molecules tend to self-assemble. After mixing the solutions, they were incubated for 20 min at room temperature (RT) and were then used in experiments (Figure S1). Depending on the experiment settings, the procedure of complex formation was slightly adjusted as noted in the particular case.
The same procedure was performed with dendrimers PPD1, PPD2, and PPD3.
Experimental Protocol for Transduction Assay
For transduction assays, 96-well plates were used containing 2 × 104 cells per well, which were seeded the day before transduction. Cells were infected with pMOI 1000 unless otherwise specified and incubated for 24 h (CAR positive cell lines such as A549 cells, Hela cells, and MDCK-2 cells) or 72 h (CAR negative cell lines such as SKOV-3 cells, CHO-K1 cells, and PTK1 cells) at 37 °C. EGFP positive cells and the overall mean fluorescence intensity (MFI) of EGFP expression was measured by flow cytometry.
PPD3/Ad5 Transduction of CAR Positive and CAR Negative Cells
Hela and MDCK-2 cells were maintained in MEM (minimum essential medium) (Gibco) supplemented with 10% heat inactivated FCS, 1% l-glutamine, and 1% penicillin/streptomycin. CHO-K1 cells were maintained in F-12 Nut Mix (HAM) 1X (Gibco) supplemented with 10% heat inactivated FCS and 1% penicillin/streptomycin. PTK1 cells were maintained in DMEM supplemented with 10% heat inactivated FCS, 1% glutamine, and 1% penicillin/streptomycin. All the cells were maintained at 37 °C under 5% CO2. Then 50 μL of dendrimers and Ad5 solutions were left to equilibrate for 1 h at 37 °C and then added to 1 mL of medium in a 12-well flat bottom plate containing 105 cells (MOI = 10–100) for 24–48 h at 37 °C with medium supplemented with 2% of FCS. The same experimental conditions were used in other cell lines. The cells were harvested 24–48 h post-transduction, fixed in 1% paraformaldehyde, stored at 4 °C in PBS, and protected from light. The fraction of infected cells was quantified by flow cytometry, which measured GFP signal of infected cells. The sample acquisitions were performed with FACSCalibur (BD Biosciences; USA), and 10 000 events were acquired for each sample. Untreated cells were set as negative control. Each experiment was performed in triplicate, and the data were analyzed from two independent experiments.
Animal Experiment and Bioluminescence Imaging
All animal studies were performed in compliance with protocols that had been approved by the Hubei Provincial Animal Care and Use Committee and the experimental guidelines of the Animal Experimentation Ethics Committee of Huazhong University of Science and Technology. Male C57BL/6J mice (purchased from China Three Gorges University) at the age of 9 weeks were used for in vivo experiments. All mice were housed in a specific pathogen-free facility. All mouse experiments were performed in compliance with institutional guidelines and according to the protocol approved by the Institutional Animal Care and Use Committee of Huazhong University of Science and Technology. The mice were divided into three groups: control group (3 mice), Ad5 group (6 mice), and Ad5+PPD3 group (6 mice). Ad5 (2 × 1010 VP) or Ad5 incubation with PPD3 (Ad5/PPD3 = 1:10 000, after 20 min) in 200 μL of PBS were administered via tail vein injection. After 2 weeks, all animals were killed. Then the hearts, livers, spleens, lungs, kidneys, and brains were harvested, and EGFP expression in the organs was visualized by bioluminescence imaging at 488 nm/507 nm (excitation/emission). After that, half of the organs were cryopreserved at −80 °C, and half were fixed with fixative (4% formaldehyde, 50% ethyl alcohol, 2% acetic acid).
Western Blot Assay
All organs were ground, and whole proteins were extracted with lysis buffer (50 mM HEPES, pH 7.4, containing 150 mM NaCl, 1% Triton X-100, 0.4% SDS, protease inhibitor cocktail). Protein concentrations were determined by the bicinchoninic acid (BCA) protein assay kit. Samples were heated to 100 °C for 10 min in loading buffer with SDS and β-mercaptoethanol, run on 12% SDS-PAGE gels in electrophoresis buffer (25 mmol/L Tris, 250 mmol/L glycine, 0.1% SDS), and transferred to PVDF membranes. All blots were blocked in 5% skim milk in TBST (100 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1% Tween20) at room temperature for 1 h. Membranes were probed with rabbit anti-EGFP, anti-GAPDH, or anti-β-actin (primary antibodies were used at a dilution of 1:4000 in 5% bovine serum albumin (BSA)–TBST). The HRP-linked secondary antibody was diluted 1:2000 in 5% milk–TBST and incubation at room temperature for 1 h. The blots were visualized using an ECL detection system.
Immunohistochemistry
Organs were fixed with formaldehyde and embedded in paraffin. For immunohistochemistry, thin sections of the embedded tissues were then blocked with 5% BSA for 2 h at room temperature and incubated with anti-EGFP (1:200) at 4 °C overnight. After washing for 15 min, the sections were incubated with biotin-conjugated secondary antibody for 30 min and further incubated with a solution of DAB. The sections were then dehydrated in ascending grades of ethyl alcohol, cleared with xylene, and covered with a coverslip.
Statistical Analysis
For statistical evaluation of the data, Microsoft Office Excel was used. The analysis included calculation of the mean value and the standard deviation. To compare, for example, different cell lines, relative values were computed. To analyze the data’s statistical significance, p-values were calculated with the TTEST-function (student’s t test) of Excel. P-values ≤ 0.05 were regarded as significant and labeled in graphs with one asterisk (∗). Data points with two asterisks indicated p-values ≤ 0.01 and three asterisks p-values ≤ 0.001. P-values of data points, which were regarded as significant, were also provided in brackets in the text where these data were analyzed and were marked with “p”. Samples of all experiments were analyzed in triplicates. In the caption of the graphs, this is referred to as sample number (n).
Acknowledgments
We thank Dirk Nettelbeck, Deutsches Krebsforschungszentrum Heidelberg, for providing material to construct Ad5/F41s. Weil and Müllen thank the Volkswagen Foundation (Project No. 88396) for financial support. The project is supported by an ERC Synergy Grant (319130-BioQ), the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under project number 316249678 - SFB 1279, and the 1000 Young Talent Program of China and the National Natural Science Foundation of China (51703073). Y.W. and L.L. thank the Analytical and Testing Center of Huazhong University of Science and Technology (HUST) and the Instrument Center at College of Life Science and Technology of HUST for instrument support. J.W. is a recipient of a DFG-funded position through the Excellence Initiative by the Graduate School Materials Science in Mainz (GSC 266).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.9b01484.
Experimental details on synthesis and characterization of dendrimers, in vitro and in vivo experiments (PDF)
Author Contributions
⬢ These authors contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Hadjidemetriou M.; Kostarelos K. Nanomedicine: Evolution of the Nanoparticle Corona. Nat. Nanotechnol. 2017, 12, 288–290. 10.1038/nnano.2017.61. [DOI] [PubMed] [Google Scholar]
- Chen F.; Wang G.; Griffin J. I.; Brenneman B.; Banda N. K.; Holers V. M.; Backos D. S.; Wu L.; Moghimi S. M.; Simberg D. Complement Proteins Bind to Nanoparticle Protein Corona and Undergo Dynamic Exchange In Vivo. Nat. Nanotechnol. 2017, 12, 387–393. 10.1038/nnano.2016.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Aberg C.; Mahon E.; Dawson K. A. Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat. Nanotechnol. 2013, 8, 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Tenzer S.; Docter D.; Kuharev J.; Musyanovych A.; Fetz V.; Hecht R.; Schlenk F.; Fischer D.; Kiouptsi K.; Reinhardt C.; Landfester K.; Schild H.; Maskos M.; Knauer S. K.; Stauber R. H. Rapid Formation of Plasma Protein Corona Critically Affects Nanoparticle Pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- Schottler S.; Becker G.; Winzen S.; Steinbach T.; Mohr K.; Landfester K.; Mailander V.; Wurm F. R. Protein Adsorption Is Required for Stealth Effect of Poly(Ethylene Glycol)- and Poly(Phosphoester)-Coated Nanocarriers. Nat. Nanotechnol. 2016, 11, 372–377. 10.1038/nnano.2015.330. [DOI] [PubMed] [Google Scholar]
- Fletcher S.; Hamilton A. D. Protein Surface Recognition and Proteomimetics: Mimics of Protein Surface Structure and Function. Curr. Opin. Chem. Biol. 2005, 9, 632–638. 10.1016/j.cbpa.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Bisker G.; Dong J.; Park H. D.; Iverson N. M.; Ahn J.; Nelson J. T.; Landry M. P.; Kruss S.; Strano M. S. Protein-Targeted Corona Phase Molecular Recognition. Nat. Commun. 2016, 7, 10241. 10.1038/ncomms10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A.; Uzun O.; Hu Y.; Hu Y.; Han H. S.; Watson N.; Chen S.; Irvine D. J.; Stellacci F. Surface-Structure-Regulated Cell-Membrane Penetration by Monolayer-Protected Nanoparticles. Nat. Mater. 2008, 7, 588–595. 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.; Qiu Q.; Tian J.; Smith J. S.; Conenello G. M.; Morita T.; Byrnes A. P. Coagulation Factor X Shields Adenovirus Type 5 From Attack by Natural Antibodies and Complement. Nat. Med. 2013, 19, 452–457. 10.1038/nm.3107. [DOI] [PubMed] [Google Scholar]
- Waddington S. N.; McVey J. H.; Bhella D.; Parker A. L.; Barker K.; Atoda H.; Pink R.; Buckley S. M.; Greig J. A.; Denby L.; Custers J.; Morita T.; Francischetti I. M.; Monteiro R. Q.; Barouch D. H.; van Rooijen N.; Napoli C.; Havenga M. J.; Nicklin S. A.; Baker A. H. Adenovirus Serotype 5 Hexon Mediates Liver Gene Transfer. Cell 2008, 132, 397–409. 10.1016/j.cell.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Waddington S. N.; Parker A. L.; Havenga M.; Nicklin S. A.; Buckley S. M.; McVey J. H.; Baker A. H. Targeting of Adenovirus Serotype 5 (Ad5) and 5/47 Pseudotyped Vectors In Vivo: Fundamental Involvement of Coagulation Factors and Redundancy of CAR Binding by Ad5. J. Virol. 2007, 81, 9568–9571. 10.1128/JVI.00663-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal R. G. Adenovirus: The First Effective In Vivo Gene Delivery Vector. Hum. Gene Ther. 2014, 25, 3–11. 10.1089/hum.2013.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos S. K.; Barry M. A. Current Advances and Future Challenges in Adenoviral Vector Biology and Targeting. Curr. Gene Ther. 2007, 7, 189–204. 10.2174/156652307780859062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayakhmetov D. M.; Gaggar A.; Ni S.; Li Z. Y.; Lieber A. Adenovirus Binding to Blood Factors Results in Liver Cell Infection and Hepatotoxicity. J. Virol. 2005, 79, 7478–7491. 10.1128/JVI.79.12.7478-7491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba R.; Bradshaw A. C.; Parker A. L.; Bhella D.; Waddington S. N.; Nicklin S. A.; van Rooijen N.; Custers J.; Goudsmit J.; Barouch D. H.; McVey J. H.; Baker A. H. Identification of Coagulation Factor (F)X Binding Sites on the Adenovirus Serotype 5 Hexon: Effect of Mutagenesis on FX Interactions and Gene Transfer. Blood 2009, 114, 965–971. 10.1182/blood-2009-03-208835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi H.; Hayakawa T. Targeted Adenovirus Vectors. Hum. Gene Ther. 2004, 15, 1034–1044. 10.1089/hum.2004.15.1034. [DOI] [PubMed] [Google Scholar]
- Fuxe J.; Liu L.; Malin S.; Philipson L.; Collins V. P.; Pettersson R. F. Expression of the Coxsackie and Adenovirus Receptor in Human Astrocytic Tumors and Xenografts. Int. J. Cancer 2003, 103, 723–729. 10.1002/ijc.10891. [DOI] [PubMed] [Google Scholar]
- Stangenberg R.; Wu Y.; Hedrich J.; Kurzbach D.; Wehner D.; Weidinger G.; Kuan S. L.; Jansen M. I.; Jelezko F.; Luhmann H. J.; Hinderberger D.; Weil T.; Mullen K. A Polyphenylene Dendrimer Drug Transporter with Precisely Positioned Amphiphilic Surface Patches. Adv. Healthcare Mater. 2015, 4, 377–384. 10.1002/adhm.201400291. [DOI] [PubMed] [Google Scholar]
- Stangenberg R.; Saeed I.; Kuan S. L.; Baumgarten M.; Weil T.; Klapper M.; Mullen K. Tuning Polarity of Polyphenylene Dendrimers by Patched Surface Amphiphilicity--Precise Control over Size, Shape, and Polarity. Macromol. Rapid Commun. 2014, 35, 152–160. 10.1002/marc.201300671. [DOI] [PubMed] [Google Scholar]
- Hammer B. A. G.; Wu Y.; Fischer S.; Liu W.; Weil T.; Mullen K. Controlling Cellular Uptake and Toxicity of Polyphenylene Dendrimers by Chemical Functionalization. ChemBioChem 2017, 18, 960–964. 10.1002/cbic.201700079. [DOI] [PubMed] [Google Scholar]
- Wiesler U. M.; Mullen K. Polyphenylene Dendrimers via Diels-Alder Reactions: The Convergent Approach. Chem. Commun. 1999, 2293–2294. 10.1039/a907339f. [DOI] [Google Scholar]
- Bergelson J. M.; Cunningham J. A.; Droguett G.; Kurt-Jones E. A.; Krithivas A.; Hong J. S.; Horwitz M. S.; Crowell R. L.; Finberg R. W. Isolation of a Common Receptor for Coxsackie B Viruses and Adenoviruses 2 and 5. Science 1997, 275, 1320–1323. 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Callipo L.; De Sanctis S. C.; Cavaliere C.; Pozzi D.; Lagana A. Surface Adsorption of Protein Corona Controls the Cell Internalization Mechanism of DC-Chol-DOPE/DNA Lipoplexes in Serum. Biochim. Biophys. Acta, Biomembr. 2010, 1798, 536–543. 10.1016/j.bbamem.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Kokkinopoulou M.; Simon J.; Landfester K.; Mailander V.; Lieberwirth I. Visualization of the Protein Corona: towards a Biomolecular Understanding of Nanoparticle-Cell-Interactions. Nanoscale 2017, 9, 8858–8870. 10.1039/C7NR02977B. [DOI] [PubMed] [Google Scholar]
- Shen L. M.; Tenzer S.; Storck W.; Hobernik D.; Raker V. K.; Fischer K.; Decker S.; Dzionek A.; Krauthauser S.; Diken M.; Nikolaev A.; Maxeiner J.; Schuster P.; Kappel C.; Verschoor A.; Schild H.; Grabbe S.; Bros M. Protein Corona-Mediated Targeting of Nanocarriers to B Cells Allows Redirection of Allergic Immune Responses. J. Allergy Clin. Immunol. 2018, 142, 1558–1570. 10.1016/j.jaci.2017.08.049. [DOI] [PubMed] [Google Scholar]
- Caracciolo G.; Cardarelli F.; Pozzi D.; Salomone F.; Maccari G.; Bardi G.; Capriotti A. L.; Cavaliere C.; Papi M.; Lagana A. Selective Targeting Capability Acquired with a Protein Corona Adsorbed on the Surface of 1,2-Dioleoyl-3-Trimethylammonium Propane/DNA Nanoparticles. ACS Appl. Mater. Interfaces 2013, 5, 13171–13179. 10.1021/am404171h. [DOI] [PubMed] [Google Scholar]
- Di Paolo N. C.; Kalyuzhniy O.; Shayakhmetov D. M. Fiber Shaft-Chimeric Adenovirus Vectors Lacking the KKTK Motif Efficiently Infect Liver Cells In Vivo. J. Virol. 2007, 81, 12249–12259. 10.1128/JVI.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe H. A.; Danel C.; Longenecker G.; Metzger M.; Setoguchi Y.; Rosenfeld M. A.; Gant T. W.; Thorgeirsson S. S.; Stratford-Perricaudet L. D.; Perricaudet M.; Pavirani A.; Lecocq J. P.; Crystal R. G. Adenovirus-Mediated In Vivo Gene Transfer and Expression in Normal Rat Liver. Nat. Genet. 1992, 1, 372–378. 10.1038/ng0892-372. [DOI] [PubMed] [Google Scholar]
- Schnell M. A.; Zhang Y.; Tazelaar J.; Gao G. P.; Yu Q. C.; Qian R.; Chen S. J.; Varnavski A. N.; LeClair C.; Raper S. E.; Wilson J. M. Activation of Innate Immunity in Nonhuman Primates Following Intraportal Administration of Adenoviral Vectors. Mol. Ther. 2001, 3, 708–722. 10.1006/mthe.2001.0330. [DOI] [PubMed] [Google Scholar]
- Tao N.; Gao G. P.; Parr M.; Johnston J.; Baradet T.; Wilson J. M.; Barsoum J.; Fawell S. E. Sequestration of Adenoviral Vector by Kupffer Cells Leads to a Nonlinear Dose Response of Transduction in Liver. Mol. Ther. 2001, 3, 28–35. 10.1006/mthe.2000.0227. [DOI] [PubMed] [Google Scholar]
- Alba R.; Bradshaw A. C.; Coughlan L.; Denby L.; McDonald R. A.; Waddington S. N.; Buckley S. M.; Greig J. A.; Parker A. L.; Miller A. M.; Wang H.; Lieber A.; van Rooijen N.; McVey J. H.; Nicklin S. A.; Baker A. H. Biodistribution and Retargeting of FX-Binding Ablated Adenovirus Serotype 5 Vectors. Blood 2010, 116, 2656–2664. 10.1182/blood-2009-12-260026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker A. L.; Waddington S. N.; Nicol C. G.; Shayakhmetov D. M.; Buckley S. M.; Denby L.; Kemball-Cook G.; Ni S.; Lieber A.; McVey J. H.; Nicklin S. A.; Baker A. H. Multiple Vitamin K-Dependent Coagulation Zymogens Promote Adenovirus-Mediated Gene Delivery to Hepatocytes. Blood 2006, 108, 2554–2561. 10.1182/blood-2006-04-008532. [DOI] [PubMed] [Google Scholar]
- Vigant F.; Descamps D.; Jullienne B.; Esselin S.; Connault E.; Opolon P.; Tordjmann T.; Vigne E.; Perricaudet M.; Benihoud K. Substitution of Hexon Hypervariable Region 5 of Adenovirus Serotype 5 Abrogates Blood Factor Binding and Limits Gene Transfer to Liver. Mol. Ther. 2008, 16, 1474–1480. 10.1038/mt.2008.132. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.