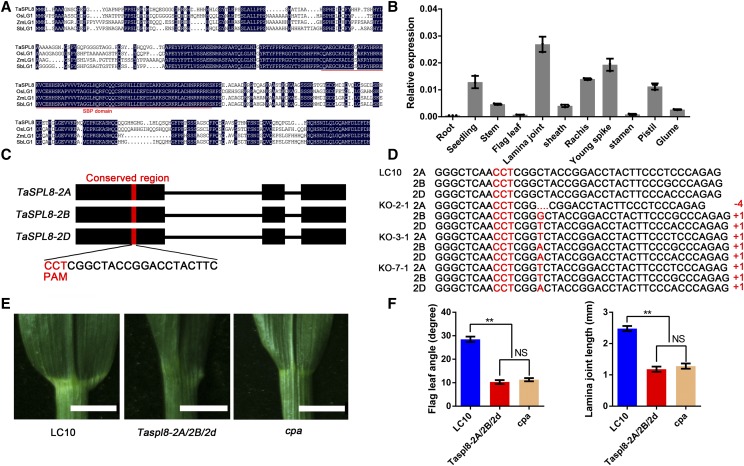
Figure 3.
Cloning and characterization of TaSPL8 and the phenotype of its knockout (KO) lines by the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein9 (Cas9) system. A, Amino acid sequence alignment of TaSPL8 and its orthologs in rice (Os04g0656500), maize (Zm00001d002005), and sorghum (SORBI_3006G247700). The conserved SQUAMOSA-PROMOTER BINDING PROTEIN domain is indicated by a red line. B, Expression pattern of TaSPL8 in various tissues. The expression levels were normalized to wheat TaACTIN. Each bar in the graph corresponds to the mean value of three data points, shown by dots. C, Sequence of a single-guide RNA (sgRNA) designed to target a conserved region of exon 1 among the three TaSPL8 homeologs. The protospacer-adjacent motif (PAM) sequence is highlighted in red. D, The genotypes of three mutant lines were identified by sequencing. The symbols “+” and “−” indicate the insertion or deletion caused by CRISPR/Cas9-induced mutations, respectively. Numbers indicate the length of the insertion or deletion. E, Flag leaf phenotypes of mutant Taspl8-2A/2B/2d, cpa and LC10. Bars = 1 cm. F, Flag leaf angle and lamina joint length of Taspl8-2A/2B/2d, cpa and LC10 plants. The double asterisk represents significant differences determined by two-tailed Student’s t test at P < 0.01. Error bars = sd (n = 10).