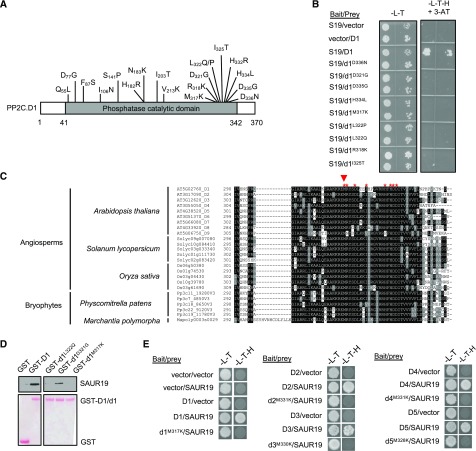
Figure 1.
Screening of SAUR interaction mutants of PP2C.D1. A, Diagram of the PP2C.D1 protein with point mutations that abolish SAUR19 binding indicated. B, Reverse yeast two-hybrid assay of wild-type PP2C.D1 or mutant pp2c.d1 derivatives with SAUR19. Growth on SC-Leu,Trp,His (-L-T-H) + 3-amino-1′, -2′, -4′ triazole (3-AT) medium is indicative of PP2C.D1-SAUR19 interaction. C, Multiple sequence alignment of partial PP2C.D amino acid sequences from a variety of plants. Not all tomato (Solanum lycopersicum) and rice (Oryza sativa) members are included. PP2C.D orthologs from other species were retrieved from https://phytozome.jgi.doe.gov/pz/portal.html. Red asterisks indicate the positions of PP2C.D1 residues identified in the reverse two-hybrid screen. The red arrowhead indicates the conserved Met residue, which was mutated to Lys in other PP2C.D family members. D, In vitro pull-down assay of SAUR19 by wild-type PP2C.D1 or mutated pp2c.d1 derivatives. Immunoblots are shown in the top row, whereas Ponceau S-stained membranes are shown in the bottom row. All samples were run on the same gel and blotted together. Extraneous lanes were digitally spliced out of the blot as indicated. E, Yeast two-hybrid assay of wild-type PP2C.D or mutant pp2c.d isoforms with SAUR19. In B and E, images are composites where individual images were cropped and digitally extracted for comparison.