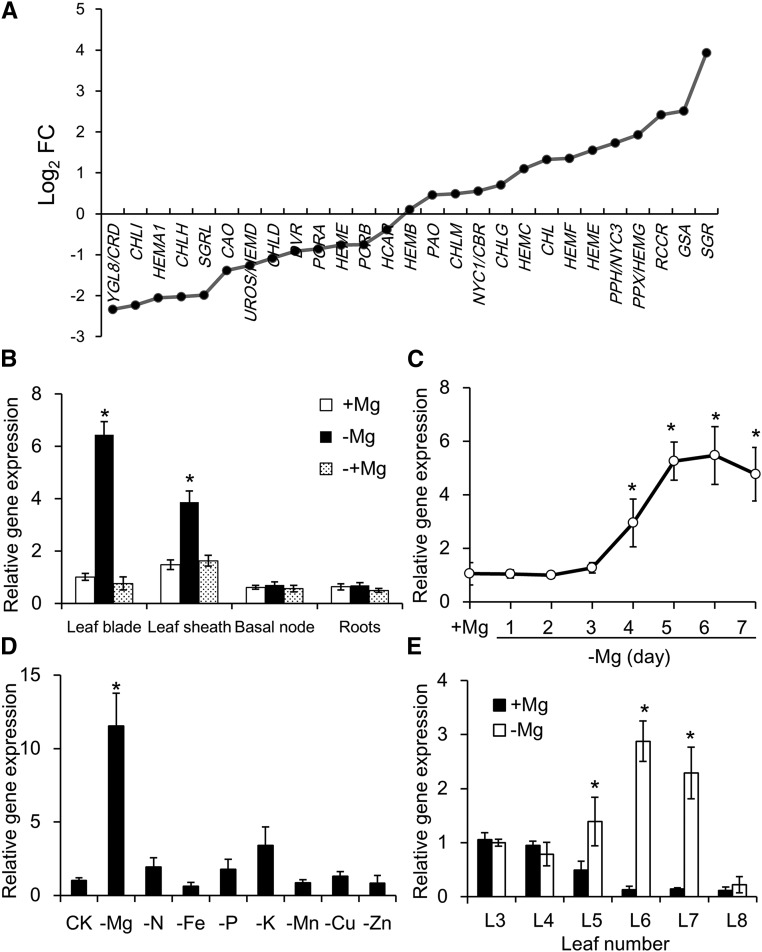
Figure 2.
Gene expression patterns in response to Mg deprivation. A, Transcriptomic analysis of genes involved in chlorophyll biosynthesis and degradation of rice shoots. Fold change (FC) of fragments per kilobase million value in −Mg-treated plants relative to +Mg-treated plants is shown. B, Tissue-specific expression of OsSGR. Rice seedlings were grown in nutrient solution containing 0- or 250-μ m Mg for 7 d. Afterward, Mg-deficient seedlings were resupplied with 250-μ m Mg for another day. Expression is shown relative to expression in +Mg leaf blades. C, Time-dependent expression of OsSGR. Three rice seedlings were put into −Mg nutrient solution each day and all were harvested 7 d after transferring the first plants. Expression is shown relative to expression in +Mg plants. D, Mg-specific response of OsSGR expression. Rice seedlings were grown in normal nutrient solution (CK) or in the nutrient solution without Mg, N, Fe, P, K, Mn, Cu, or Zn for 7 d. Expression is shown relative to expression in CK plants. E, Expression of OsSGR in different leaves. L3–L8 leaves were separated and harvested after growing plants in nutrient solution containing 0- or 250-μ m Mg for 7 d. Expression is shown relative to expression in +Mg L3 leaves. The expression level was determined by quantitative reverse transcription PCR. Data are means ± sd (n = 3). The asterisk shows a significant difference compared to +Mg treatment (P < 0.05 by Tukey’s test).