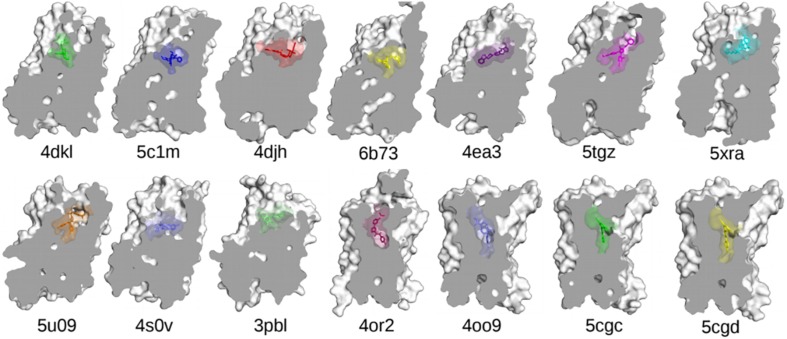
FIGURE 3.
The experimentally determined high-resolution GPCR structures, together with their bound ligands, used in the MD-based studies discussed in this review. Nanobodies and other interacting proteins were removed. PDB 4dkl, The antagonist β-FNA bound to the MOP receptor; PDB 5c1m, The agonist BU72 bound to the MOP receptor; PDB 4djh, The antagonist JDTic bound to the KOP receptor; PDB 6b73, The agonist MP1104 bound to the KOP receptor; PDB 4ea3, The peptide mimetic antagonist compound 24 bound to the NOP receptor; PDB 5tgz, The antagonist AM6538 bound to the CB1 receptor; PDB 5xra, The agonist AM11542 bound to the CB1 receptor; PDB 5u09, The inverse agonist taranabant bound to the CB1 receptor; PDB 4s0v, The antagonist suvorexant bound to the OX2 receptor; PDB 3pbl, The antagonist eticlopride bound to the D3 receptor; PDB 4or2, The negative allosteric modulator FITM bound to the transmembrane domain of mGluR1; PDB 4oo9, The negative allosteric modulator mavoglurant bound to the transmembrane domain of mGluR5; PDB 5cgc, The negative allosteric modulator 3-chloro-4-fluoro-5-[6-(1H-pyrazol-1-yl)pyrimidin-4-yl]benzonitrile bound to the transmembrane domain of mGluR5; PDB 5cgd, The negative allosteric modulator 3-chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile bound to the transmembrane domain of mGluR5.