Figure 6.
Phylogenetic Placement of 1,132 Metagenomically Reconstructed Genomes and Mother-to-Infant Transmission of Taxonomically Uncharacterized Strains
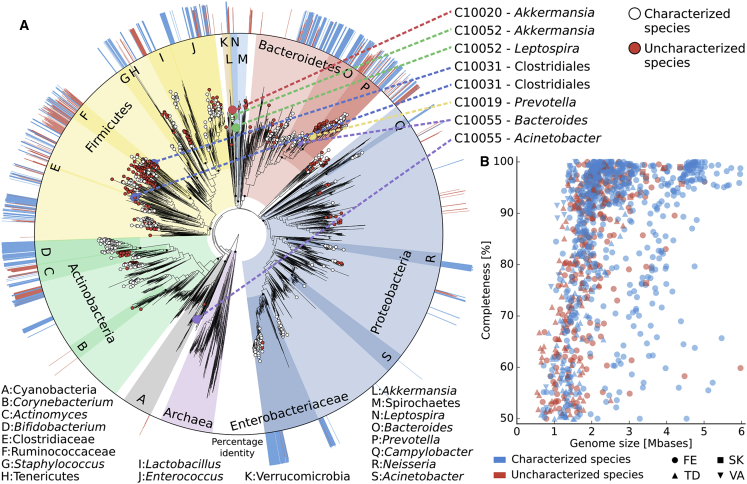
(A) We used PhyloPhlAn2 (Segata et al., 2013) to place the genomes reconstructed with metaSPAdes (Nurk et al., 2017) and binned with MetaBAT2 (Kang et al., 2015) (STAR Methods) on the microbial “Tree of Life” (Ciccarelli et al., 2006, Segata et al., 2013), which encompasses 4,000 species with available reference genomes. Leaf nodes without circles refer to reference genomes from known species, white circles indicate reconstructed genomes that are close (>95% identity) to a known species, and red circles show reconstructed genomes that cannot be assigned (<95% identity) to known species. The eight events of mother-to-infant transmission of strains from species yet to be described are called out on the top right, and the external ring of the phylogeny reports the percent identity of each leaf node against the closest genomes from known species (values below 95% are shown in red).
(B) The reconstructed genomes with completeness >50% from each body site are plotted with the corresponding completeness and genome size.