FIG 5.
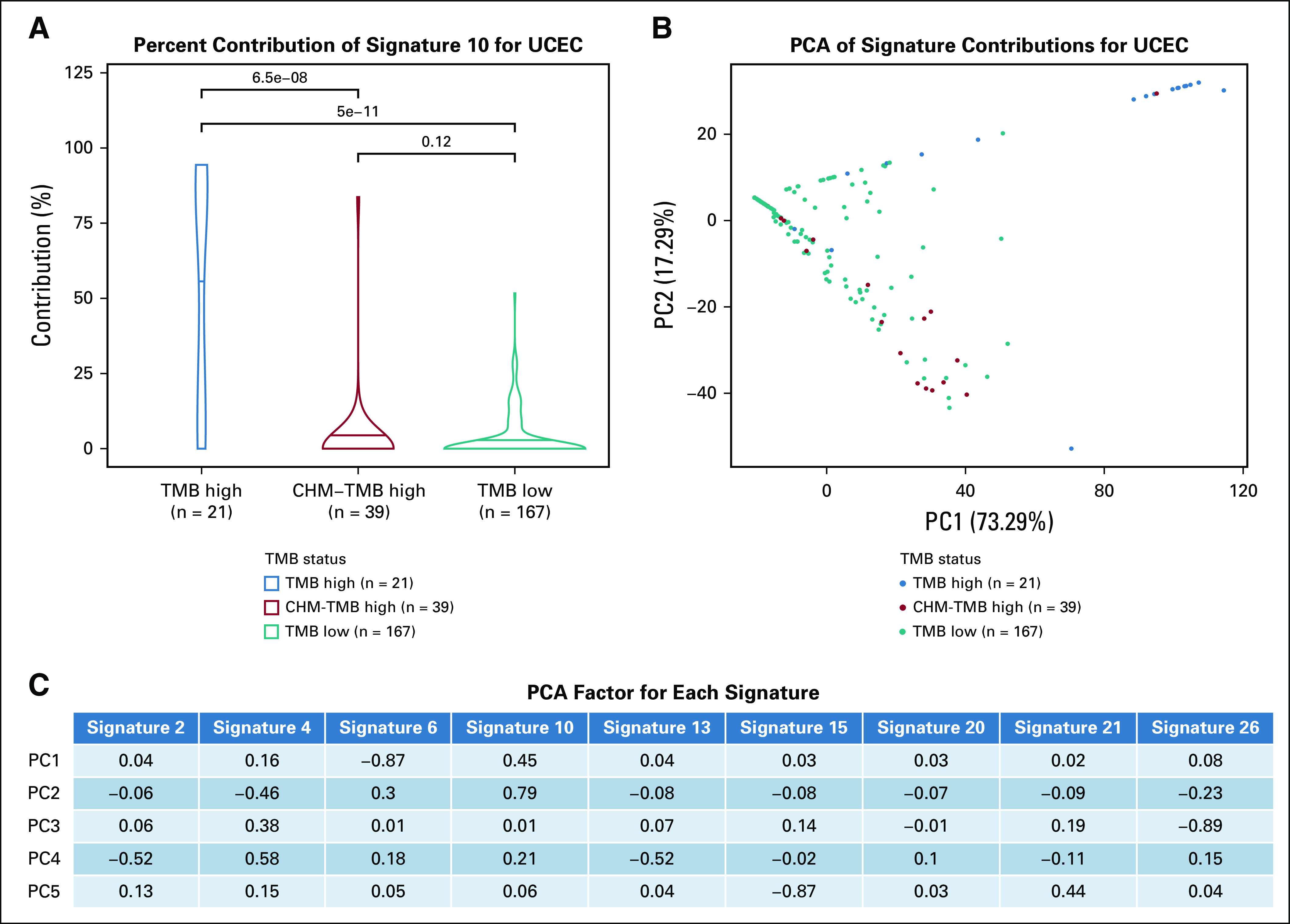
Signature contributions for uterine corpus endometrial carcinoma (UCEC) show that samples cluster by signature 10. (A) Violin plots showing the distribution of the contribution of signature 10 (defective polymerase-ε) in tumor mutational burden (TMB) high, Chalmers (CHM)-TMB high, and TMB low. The mean contribution is significantly different between TMB high and TMB low (Wilcoxon P value = 5e-11), and between TMB high and CHM-TMB high (Wilcoxon P value = 6.5e-8). (B) PCA showing the signature contributions for uterine corpus endemetrial carcinoma (UCEC) with TMB-high samples in blue, WCM-TMB–high samples in red, and TMB-low samples in teal. Only signatures 2, 4, 6, 10, 13, 15, 20, 21, and 26 were considered, to focus on signatures with the most relevant interpretations. (C) The calculated PCA factor loadings are also shown for each signature in the first five principal components. Signature 6 (defective DNA mismatch repair) has the largest factors for principal component 1 (PC1), although it is negative. PCA, principal component analysis; PC2, principal component 2.
