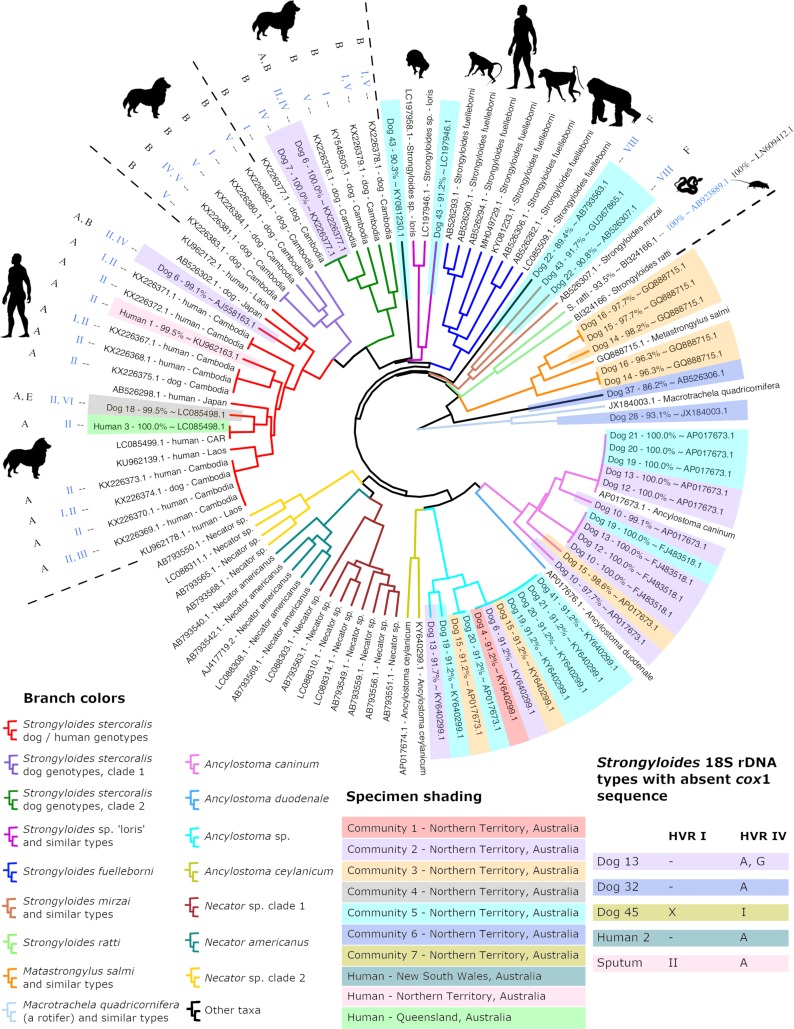
Fig 1. Dendrogram of clustered cox1 amplicons from Australian dog and human specimens.
This dendrogram includes cox1 sequences generated in this study and a selection of previously published cox1 sequences that overlap with our 217 base pair cox1 amplicon by 100%. Specimens analyzed as part of this study are shaded according to their site of collection. Branches are color coded according to their identity; either a species assignment, a proposed genus assignment, or their S. stercoralis genotype. When available, Strongyloides sp. cox1 sequences are annotated with their associated SSU haplotypes, with their HVR-I type shown in blue and their HVR-IV type shown in black. Specimens for which a cox1 sequence was not obtained are shown in a table embedded in the figure (bottom right), which includes two specimens possessing unique 18S haplotypes; dog 13 (HVR-IV, type G) and dog 45 (HVR-I, type X). A dash (-) shown in this table indicates failed amplification and/or sequencing of that marker. ‘Sputum’ refers to the sole sputum sample from a human patient (human 4) included in this study. Sequences published in previous studies that are not from S. stercoralis are labelled with their GenBank and/or DNA Data Bank of Japan accession numbers followed by their species name. Strongyloides stercoralis sequences from previous studies are labelled with their accession number, host species, and country of origin. Note that ‘CAR’ means Central African Republic. Names of specimens collected as part of this study begin with a host name and a unique number assigned in this study, followed by a percentage similarity to (~) a near BLASTN hit identifiable by its accession number.