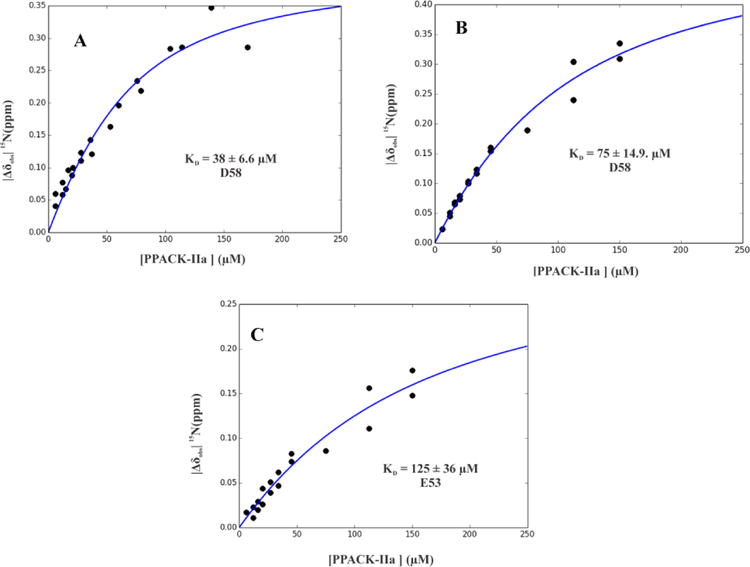
Figure 5:
Calculating affinity (KD) for 15N-labeled E53 and D58 of PAR1GED (49–62) binding to PPACK-IIa and GpIbα3P – PPACK-IIa. KD values for PAR1G D58 went from A) 38 ± 6.6 μM with PPACK-IIa to B) 75 ± 14.9 μM with GpIbα3P – PPACK-IIa. KD values for PAR1G E53 went from too weak to calculate by NMR to C) 125 ± 36 μM GpIbα3P – PPACK-IIa. NMR titrations were carried out in duplicate. The KD values were assessed from scripts written in-house through Python. A Monte-Carlo approach that assumes a 10% error in the serially diluted protein samples was employed for error analysis.