Abstract
The poly(cystaminebis(acrylamide)-diaminohexane) (poly(CBA-DAH)) was designed previously as a bio-reducible efficient gene delivery carrier. However, the high weight ratio required to form the polyplexes between poly(CBA-DAH) with pDNA is still a problem that needs to be addressed. To solve this problem and increase the transfection efficiency, poly(ethylenimine) (PEI, 1.8 kDa) was conjugated to poly(CBA-DAH) via disulfide bond. The PEI conjugated poly(CBA-DAH) (PCDP) can bind with pDNA at a very low weight ratio of 0.5 and above, like PEI 25 kDa, and form the polyplexes with nano-size (102 ~ 128 nm) and positive surface charge (27 ~ 34 mV). PCDP and PCDP polyplexes had negligible cytotoxicity and indicated similar or better cellular uptake than the comparison groups such as PEI 25 kDa and Lipofectamine® polyplexes. To confirm the transfection efficiency, the plasmid DNA (pDNA) encoded with the luciferase reporter gene (gWiz-Luc) and green fluorescent protein reporter gene (GFP) were used and treated with PCDP into the A549, Huh-7, and Mia PaCa-2 cells. PCDP/pDNA polyplexes showed highest transfection efficiency in all tested cell lines. In the luciferase assay, PCDP polyplexes showed 10.2 times higher gene transfection efficiency than Lipofectamine® polyplexes in mimic in vivo conditions (30% FBS, A549 cells). The VEGF siRNA expressing plasmid (pshVEGF), which is constructed as a therapeutic gene by our previous work, was delivered by PCDP into the cancer cells. The VEGF gene expression of PCDP/pshVEGF polyplexes was dramatically lower than control and the VEGF gene silencing efficiencies of PCDP/pshVEGF (w/w; 10/1) polyplexes were 54% (A549 cells), 77% (Huh-7 cells), and 66% (Mia PaCa-2 cells). In addition, PCDP/pshVEGF had reduced cell viability rates of about 31% (A549 cells), 39% (Huh-7 cells), and 42% (Mia PaCa-2 cells) and showed better results than all comparison groups. In the transfection efficiency and VEGF silencing assay, PCDP polyplexes showed better results than poly(CBA-DAH) at 4-fold lower weight ratio. The data of all experiments demonstrate that the synthesized PCDP could be used for efficient gene delivery and could be widely applied.
Keywords: Bio-reducible polymer, VEGF, Disulfide bond, Gene delivery
Graphical abstract
1. INTRODUCTION
Gene therapy is the administration of genetic materials (plasmid deoxyribonucleic acid (pDNA), small interfering ribonucleic acid (siRNA), and messenger RNA (mRNA), etc.) into specific cells for the treatment of genetic and acquired disorders (Niidome & Huang, 2002; Stone, 2010). In the early days, gene therapy only focused on inherited genetic disorders, but it has recently been applied in various disorders including different forms of cancers, vascular diseases, some autosomal dominant disorders, emphysema, retinitis pigmentosa, diabetes, hemophilia, and neurodegenerative disorders (Katz, Fargnoli, Williams, & Bridges, 2013; Nayerossadat, Maedeh, & Ali, 2012). However, there are limitations in delivering only the genetic materials itself. Genetic materials have low cellular uptake efficiency due to their hydrophilic property, large size, high anionic charge density, and susceptibility toward nuclease-mediated degradation (Al-Dosari & Gao, 2009; Layek & Singh, 2013). Therefore, the carriers (commonly called vectors) have been used to solve the problem without any degradation of genetic materials.
Viral vectors such as adenovirus, lentivirus, retrovirus, adeno-associated virus, and herpes simplex virus served as vehicles for efficient gene transfer. However, viral vectors have serious concerns including inflammation, difficulty in production, inflammatory response, immunogenicity, and carcinogenicity (Al-Dosari & Gao, 2009; Bouard, Alazard-Dany, & Cosset, 2009; Layek & Singh, 2013; Yin et al., 2014). Non-viral vectors such as liposomes, micelles, polycationic polymer, and polyanionic polymer are alternatives that addresses the concerns of viral vectors. Non-viral vectors have lower gene transfection efficiency than viral vectors, but are less toxic and immunogenic than viral vectors (Al-Dosari & Gao, 2009; Eroglu et al., 2013; Xu, Wang, & Pack, 2010; Yin et al., 2014). In addition, non-viral vectors have some advantages such as the potential for repeated administration and ease of modification and production.
Over the past decade, our group synthesized various types of polycationic polymers are evaluated their application as a genetic materials delivery carrier (T. I. Kim, Ou, Lee, & Kim, 2009; H. Y. Nam, Nam, Lee, Kim, & Bull, 2012; J. P. Nam, Nam, Nah, & Kim, 2015; K. Nam, Jung, Nam, & Kim, 2015; Ou et al., 2008). To enhance transfection efficiency, these polycationic polymers required important properties such as the ability to form nano-sized polyplexes with genetic materials (T. I. Kim et al., 2009; H. Y. Nam et al., 2012; J. P. Nam et al., 2015; K. Nam et al., 2015; Ou et al., 2008), bio-reducible property (T. I. Kim et al., 2009; H. Y. Nam et al., 2012; J. P. Nam et al., 2015; K. Nam et al., 2015; Ou et al., 2008), cell penetrating property (T. I. Kim et al., 2009; J. P. Nam et al., 2015), and endosome escape property (H. Y. Nam et al., 2012; J. P. Nam et al., 2015; K. Nam et al., 2015). These polycationic polymers showed enhanced transfection efficiency, however, the problems that needs to be solved in order to develop a more effective gene carrier still remain. These problems include higher weight ratio above 40 that is optimized ratio to form formation nano-sized polyplexes (T. I. Kim et al., 2009; J. P. Nam et al., 2015; Ou et al., 2008), but limits its use in vivo, and complex synthesis steps (H. Y. Nam et al., 2012; K. Nam et al., 2015).
Here, we synthesized poly(ethylenimine) (PEI) conjugated poly(cystaminebis(acrylamide)-diaminohexane)) (poly(CBA-DAH)) (PCDP) to decrease weight ratio and increase transfection efficiency. We hypothesized that the PEI provide increased binding ability and enhanced endosome escape property to poly(CBA-DAH), leading to formation of polyplexes with low weight ratios and high transfection efficiency. In addition, PEI was conjugated to poly(CBA-DAH) through the disulfide bond. Poly(CBA-DAH) is composed of multiple disulfide bonds, where these disulfide bonds can be cleaved in the cytoplasm by an intracellular reducing agent such as glutathione (GSH) (Doss, Debottam, & Debajyoti, 2013; Hong et al., 2006; Ou et al., 2008; Oupicky & Li, 2014; Wen et al., 2011). GSH is composed tri-peptide and synthesized in the cytosol from precursor amino acids (Chakravarthi, Jessop, & Bulleid, 2006). The intracellular concentration of GSH (1–10 mM) is extensively higher than extracellular levels (2 µM in plasma) (Hassan & Rechnitz, 1986; Hong et al., 2006). These different intracellular or extracellular concentrations of GSH lead to selective intracellular release of genetic materials from the polycationic polymer, which also contains disulfide bonds (Hong et al., 2006; Oupicky & Li, 2014).
In this study, to confirm the successful synthesis and polyplexes formation with pDNA, PCDP was characterized using the luciferase reporter gene (gWiz-Luc) or the green fluorescent protein (gWiz-GFP). Moreover, vascular endothelial growth factor (VEGF) siRNA expressing plasmid (pshVEGF) as a therapeutic gene was used to investigate the therapeutic efficiency in various cancer cells.
2. EXPERIMENTAL SECTION
2.1. Materials
gWiz-Luc and gWiz-GFP were amplified in E.coli DH5α and isolated by NucleoBond® Xtra Maxi Plus EF kit (MACHEREY-NAGEL GmbH & Co. KG, Dűren, Germany). The luciferase assay kit, agarose, and 5X reporter lysis buffer were purchased from Promega (Madison, WI). pshVEGF was designed and conducted according to our previous work (H. A. Kim, Nam, & Kim, 2014). tert-Butyl-N-(6-aminohexyl) carbamate (N-Boc-1,6-diaminohexane, N-Boc-DAH), 3- [4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT), 1,4-dithiothreitol (DTT), trifluoroacetic acid (TFA), piperidine, N,N-diisopropylethylamine (DIPEA), triisopropylsilane (TIS), dimethylformamide (DMF), dimethyl sulfoxide (DMSO), methanol (MeOH), and trypan blue solution (0.4%) were purchased from Sigma-Aldrich (St. Louis, MO). Fetal bovine serum (FBS) was purchased from Seradigm (Radnor, PA). Branched poly(ethylenimine) (bPEI, Mw = 1.8 kDa and 25 kDa), N,N’-Cystaminebisacrylamide (CBA) was purchased from PolySciences, Inc. (Warrington, PA). Spectrapor dialysis membrane was purchased from Spectrum Laboratories, Inc. (Rancho Dominguez, CA). Dulbecco’s modified Eagle’s medium (DMEM), human VEGF ELISA kit, YOYO-1 iodide (1 mM solution in DMSO), Lipofectamine® 2000 (DNA transfection reagent, 1 mg/mL), Opti-MEM® medium, trypsin-like enzyme (TrypLE™ Express), Dulbecco’s phosphate buffered saline (DPBS), and propidium iodide (PI, 1 mg/mL in H2O) were purchased from Invitrogen (Carlsbad, CA). Ellman’s reagent, Traut's reagent, succinimidyl 3-(2-pyridyldithio) propionate) (SPDP) cross linker and BCA assay kit were purchased from pierce (Rockford, IL).
2.2. Synthesis of the PEI conjugated Poly(CBA-DAH) (PCDP)
Poly(CBA-DAH), the backbone of PCDP (poly(disulfide amine)), was synthesized as per our previous work (Ou et al., 2008). To introduce the sulfhydryl group, purified poly(CBA-DAH) was dissolved in a non-amine buffer (0.1 M sodium phosphate buffer), then 4.4 equiv. of Traut’s reagent was added to the polymer in solution for 4 h at room temperature. After 4 h, the reacted solution was dialyzed against ultrapure water using a dialysis membrane (dialysis membrane (MWCO = 3,000)) to remove unreacted Traut’s reagent. Then, the product (poly(CBA-DAH)-SH) was filtered and lyophilized. The produced sulfydryl groups were measured using Ellman’s reagent.
To react between the activated NHS ester of SPDP and amine of PEI, PEI (Mw; 1.8 kDa) was dissolved in 0.1 M phosphate buffer. 1.2 equiv. of SPDP dissolved in DMF and then added to PEI solution (final solvent ratio; 1:9 (DMF:0.1 M phosphate buffer)) for 4 h at room temperature. The product (PEI-SPDP) was dialyzed against ultrapure water using a dialysis membrane (dialysis membrane (MWCO = 1,000)) and lyophilized.
Eight equiv. of PEI-SPDP was added to a prepared poly(CBA-DAH)-SH solution in 50 mM phosphate buffered saline and the mixture was stirred for 12 h at room temperature. The released pyridine-2-thione (leaving group) was monitored by UV spectroscopy at 343 nm to confirm the reaction (Carlsson, Drevin, & Axen, 1978). The final product (PCDP) was dialyzed against ultrapure water using a dialysis membrane (dialysis membrane (MWCO = 10,000)) and was filtered, followed by lyophilization. The synthesis of PCDP was estimated by measuring 1H NMR (Bruker, 400 MHz, D2O).
2.3. Acid-base titration
The buffering capacity of PCDP was measured by acid-base titration. 10 mg of each poly(CBA-DAH) and PCDP were dissolved in 10 mL of H2O (1 mg/mL). The solutions were adjusted to pH = 10 by 0.1 M NaOH and then titrated to pH = 3 with 0.01 M HCl.
2.4. Characterization of PCDP/pDNA polyplexes
PCDP was dissolved in HEPES buffer (10 mM HEPES, 1 mM NaCl, pH 7.4) at 10 mg/mL and diluted (1 mg/mL or 5 mg/mL) to form polyplexes, then mixed at a certain weight ratio ranging from 0.05 to 5 based on 300 ng of pDNA (gWiz-Luc). The mixtures were then incubated at room temperature for 30 min before use. Gel retardation assay was used to evaluate the pDNA condensation ability of PCDP. After 30 min, the incubated mixtures were loaded on a 0.8% agarose gel with PI-included loading dye (5% in 10X loading dye, v/v), followed by electrophoresis in TAE buffer (10 mM tris/HCl, 1% v/v acetic acid, 1 mM EDTA) at 100V for 40 min (J. P. Nam et al., 2015). Naked pDNA was used as a control. The migration of PI-stained pDNA in the agrarose gel was visualized by an UV illuminator (Gel Documentation System, Bio-Rad, Hercules, CA). In addition, PEI 25 kDa and poly(CBA-DAH) were used as comparison groups with same (PEI 25 kDa, w/w, 0.05 to 5) or different (poly(CBA-DAH), w/w, 1 to 20) conditions.
The particle size and zeta-potential of PCDP/pDNA polyplexes were measured by dynamic light scattering (DLS) and laser Doppler velocimetry (LDV), respectively by a Nano ZS (ZEN3600, Malvern Instruments, UK). The polyplexes were prepared in HEPES buffer (10 mM HEPES, 1 mM NaCl, pH 7.4) at various weight ratios ranging from 0.05 to 5 based on 4 µg of pDNA (gWiz-Luc). After incubation time (30 min), the polyplexes solutions were diluted using distilled water to 600 µL before measurement. In addition, to confirm the dissociation of polyplexes by a reducing agent such as DTT, the particle size and zeta-potential of PCMD/pDNA polyplexes (weight ratio 10) were measured with or without 5 mM DTT. PEI was used as a comparison group (weight ratio 1).
2.5. Cell culture and cytotoxicity assay
Human lung adenocarcinoma epithelial (A549), human hepatocellular carcinoma (Huh-7), and human pancreatic carcinoma (Mia PaCa-2) cell lines were selected and obtained from American Type Culture Collection (ATCC) (Manassas, VA). The cells were cultured in high glucose DMEM supplemented with heat-inactivated FBS (10%, v/v), without antibiotic agent at 37°C in a humidified atmosphere containing CO2 (5%, v/v).
The cytotoxicity of PCDP and PCDP/pDNA polyplexes were evaluated in cultured cells assessing cell killing using the MTT assay. When the A549, Huh-7, and Mia PaCa-2 cells reached 70 ~ 80 %, the cells were harvested by trypsinization and centrifugation, then seeded in a 24-well plate at a density of × 104 cells/well in full DMEM medium. After 24 h, the culture medium was replaced by serum-free DMEM, then the cells were treated with polymers (PCDP and PEI 25 kDa) or polyplexes (PCDP, PEI 25 kDa, poly(CBA-DAH), and Lipofectamine® polyplexes with pDNA (gWiz-Luc)). The concentration range of the treated polymers (PCDP and PEI 25 kDa) was 1 ~ 20 µg/mL. In addition, PCDP/pDNA polyplexes were prepared at various weight ratios ranging from 1 to 20, based on pDNA (gWiz-Luc, 500 ng). Poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® were used as comparison groups and formed polyplexes with pDNA at 1:40 (optimized ratio by previously work (Ou et al., 2008)), 1:1 (optimized ratio by previously work (T. I. Kim et al., 2009; J. P. Nam et al., 2015)), and 1:2 (manufacture’s recommended ratio) weight ratio, respectively. After 4 h, the medium was replaced by serum-containing DMEM (10% FBS, without antibiotic agent) and the cells were further incubated for 48 h. 30 µL stock solution of MTT (2 mg/mL in PBS) was added to each well, and the plates were incubated for another 2 h. The medium and unreacted MTT were removed by aspiration, then 300 µL of DMSO was added to each well to dissolve the formazan crystals formed in live cells. The absorbance of each well was measured at 570 nm using a microplate reader (Model 680, Bio-Rad Laboratory, Hercules, CA), and the relative cell viability (%) calculated as: Cell viability (%) = ((OD sample) - (OD blank)) / ((OD control) – (OD blank)) × 100%.
2.6. Cellular uptake study
To confirm the cellular uptake of PCDP/pDNA polyplexes, A549, Huh-7, and Mia PaCa-2 cells were seeded at a density of 1 × 105 cells/well in a 12-well plate in DMEM (10% FBS) and incubated for 24 h before cellular uptake. The medium was replaced by serum-free DMEM, then the cells were treated with polyplexes. The polyplexes of PCDP, poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® were prepared with pDNA (YOYO-1 stained gWiz-Luc, 1 molecule of the dye per 50 bp of nucleotide, 1 µg) using the same protocol as the cytotoxicity experiments. After 4 h, the medium was removed and washed with PBS. The cells were collected by trypsinization and centrifugation (3 min and 3,000 rpm). The degree of cellular uptake was investigated by a BD FACscan analyzer at a minimum of 1 × 104 cells gated per sample. The data was analyzed using De NoVo FCS Express 5 Plus software.
2.7. GFP and Luciferase expression assay
A549, Huh-7, and Mia PaCa-2 cells were seeded at a density of 5 × 104 cells/well in a 24-well plate in DMEM (10% FBS) and incubated for 24 h. To confirm the luciferase and GFP, the both gWiz-Luc and gWiz-GFP were used. pDNA (gWiz-Luc or gWiz-GFP) was complexed to form the polyplexes with the PCDP, poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® at various weight ratio described above, respectively. The prepared polyplexes were treated using the same protocol as the cytotoxicity experiments. In addition, to create an in vitro environment that mimics in vivo conditions more accurately, cells were treated with polyplexes (with gWiz-Luc) in 30% FBS.
The GFP expression was visualized after 48 h using an EVOS microscope (AMG, Bothell, WA) and each of the GFP expression levels were quantified by measuring the absorbance at 485 nm for excitation and 535 nm for emission on a Tecan Infinite M200 Pro.
For the luciferase analysis, the medium was removed after 48 h, and the cells were washed with DPBS 3 times. Then, the cells were lysed by 1× reporter lysis buffer and collected by scraping and centrifugation. Luciferase quantification was analyzed using a Tecan Infinite M200 Pro (Tecan Group Ltd., Männedorf, Switzerland). The amount of protein in the cell lysate was measured using a Pierce® BCA protein assay kit (Pierce®, Rockford, IL) according to the manufacturer’s protocol.
2.8. VEGF silencing and cancer cell growth inhibition of PCDP polyplexes
VEGF silencing efficacy of PCDP polyplexes was evaluated by ELISA using VEGF siRNA expressing plasmid (pshVEGF). A549, Huh-7, and Mia PaCa-2 cells were seeded at a density of 5 × 104 cells/well in a 24-well plate, then the PCDP/pshVEGF polyplexes were treated as described above. Poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® were also used as comparison groups. After incubation time (48 h), the medium of each well were harvested and the VEGF expression was measured by human VEGF ELISA kit according to the manufacturer’s protocol. In addition, to confirm the cell growth inhibition, the cells were treated with PCDP/pshVEGF polyplexes at weight ratios 10 and 15 as described above and cell viability was investigated by MTT analysis.(Mosmann, 1983) Cells were also transfected with polyplexes using gWiz-Luc at the same conditions, and the cell viability was investigated by MTT analysis for comparison purposes.
2.9. Statistical analysis
The all data were expressed as mean ± standard deviation (SD). Statistical significances were performed by one-way analysis of variance (ANOVA) using SPSS Statistics 17 (SPSS Inc.). The criterion for statistical significance was *P < 0.05.
3. RESULTS AND DISCUSSION
3.1. Synthesis and characterization of PCDP
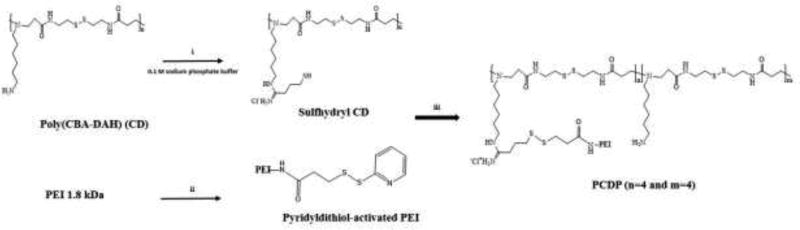
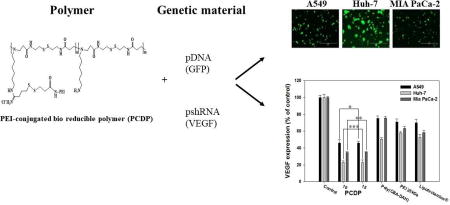
To increase the transfection efficiency and decrease the weight ratio when the polyplexes formed with pDNA, we designed a novel bio-reducible polymer (PCDP) with poly(CBA-DAH) and PEI 1.8 kDa. PEI can introduce high binding ability with pDNA as well as enhanced endosome escape ability of pDNA from the polyplexes, leading to decreased weight ratio and increased transfection efficiency (Fischer, Bieber, Li, Elsasser, & Kissel, 1999; Wang et al., 2016). Scheme 1 shows the synthetic route of PCDP.
Scheme 1.
Synthesis scheme and molecular structure of PCDP. i) Traut’s reagent in 0.1 M sodium phosphate buffer, RT, 4 h, ii) SPDP in DMF:0.1 M sodium phosphate buffer co-solvent (1:9), RT, 4 h, iii) 50 mM phosphate buffer, RT, 12 h.
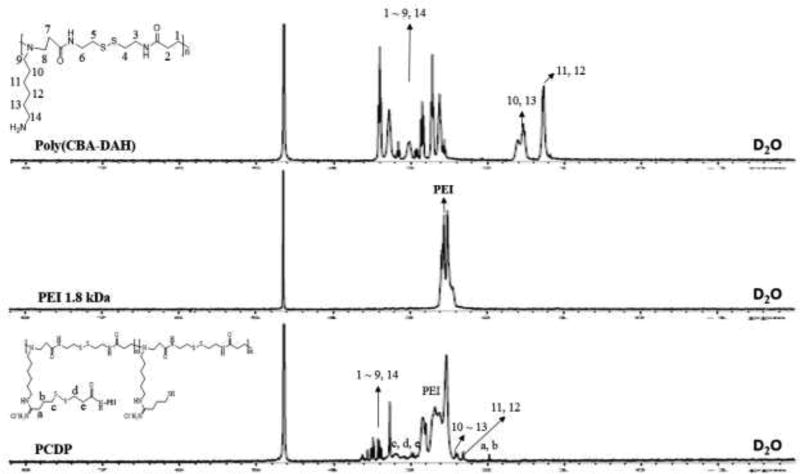
To introduce the disulfide bond between poly(CBA-DAH) and PEI 1.8 kDa, Traut’s reagent and SPDP were used. The conjugation ratio of sulfydryl group was measured by Ellman’s reagent and each reaction step was monitored by Thin-Layer Chromatography (TLC). In the first attempt, we designed PCDPs with different conjugation ratios of PEI such as 1, 4, and 8, to compare the effect of the conjugation ratio. However, in the case of conjugation ratio of 1, there was not a significant difference when compared to poly(CBA-DAH), and in the case of conjugation ratio of 8, it aggregated during the final synthesis step (data not shown). Therefore, we used PCDP with conjugation ratio of 4 for experiments to confirm its potential as a gene delivery carrier. The synthesis and conjugation ratio of PCPD were confirmed by 1H NMR. The poly(CBA-DAH) that was used as a backbone was characterized in our previous work (Ou et al., 2008). As shown in Figure 1, the peak assignments of poly(CBA-DAH), PEI 1.8 kDa, and PCDP were classified in the data. In the NMR data of PCDP, the proton peaks of poly(CBA-DAH) and PEI were shifted downfield due to steric hindrance caused by the conjugation between poly(CBA-DAH) and PEI (J. P. Nam et al., 2014).
Figure 1.
1H NMR spectra of PCDP, PEI 1.8 kDa, and poly(CBA-DAH) in NMR solvent (D2O, 3 mg/mL).
In addition, the conjugation ratio of PEI to the poly(CBA-DAH) was calculated by the ratio of the integration of the proton spectrum peaks in the poly(CBA-DAH) (−NCH2CH2CH2CH2CH2CH2NH2) and CH2 of PEI. The calculated conjugation ratio by 1H NMR analysis is shown in Table 1. The 1H NMR results showed that the PCDP were successfully synthesized. The occurrence of PCDP spectrum peaks were classified as follows; PCDP (1H NMR, D2O): poly(CBA-DAH) (NCH2CH2CH2CH2CH2CH2NH2) = 1.16 ppm (shifted to 2.25 ppm), poly(CBA-DAH) (NCH2CH2CH2CH2CH2CH2NH2) = 1.48 ppm (shifted to 2.4 ppm), poly(CBA-DAH) (NCH2CH2CONH), (CH2SSCH2), (NCH2CH2CH2CH2CH2CH2NH2), (NCH2CH2CONH, NCH2CH2CH2CH2CH2CH2NH2), and (NCH2CH2CONHCH2) = 2.57 ~ 3.43 ppm (shifted to 3.15 ~ 3.62 ppm), PEI (NCH2CH2N) = 2.39 ~ 2.61 ppm (shifted to 2.41 ~ 2.85), cross-linker (N=CCH2CH2CH2S) = 1.98 ppm, cross-linker (N=CCH2CH2CH2S) and (SCH2CH2C=O) = 2.94 ~ 3.21 ppm.
Table 1.
Characterization of PCDP
| Conjugation ratio (%)
|
Buffer Capacityb (%) | ||
|---|---|---|---|
| expected | calca | ||
| Poly (CBA-DAH) | - | - | 55 (Ou et al., 2008) |
| PCDP | 50 | 50 | 67 |
Calculated PEI conjugation ratio of PCDP by 1H NMR analysis.
Calculated by base-acid profiles.
3.2. Buffering capacity of PCDP
PEI has many nitrogen atoms including primary, secondary, and tertiary amine groups. These amine groups can enhance the buffering capacity and cause the subsequent endosomal or lysosomal rupture and escape into the cytoplasm via a “proton sponge effect” (Benjaminsen, Mattebjerg, Henriksen, Moghimi, & Andresen, 2013; Luu et al., 2012; Wang et al., 2016). Therefore, PEI can provide enhanced buffering capacity to the PCDP, leading to increased endosomal escape into cytoplasm, as described above. The studies have been published on polymers which conjugated or modified with substances that can increase the buffering capacity such as PEI, imidazole, and histidine. These studies shown the enhanced buffering capacity and increased transfection efficacy (Bello Roufai & Midoux, 2001; Pack, Putnam, & Langer, 2000; Pires et al., 2011; Yang et al., 2008; Zhang, Duan, Wang, & Bian, 2015).
The buffering capacity of PCDP in the pH range of 10 to 3 was measured by acid-base titration assay and was calculated from the acid-base titration curve (data not shown) in the endosomal pH range (from pH7.4 to 5.1) according to the following equation;
Poly(CBA-DAH) displayed 55% protonation in a previous study (Ou et al., 2008). PCDP showed increased buffering capacity more than poly(CBA-DAH), as shown in Table 1. Because more amine groups were introduced to poly(CBA-DAH), where the additional amine groups are attributed to the conjugation of PEI. This result supported that the enhanced buffering capacity may help endosomal escape of the polyplexes.
3.3. Physicochemical characterization of PCPD/pDNA polyplexes
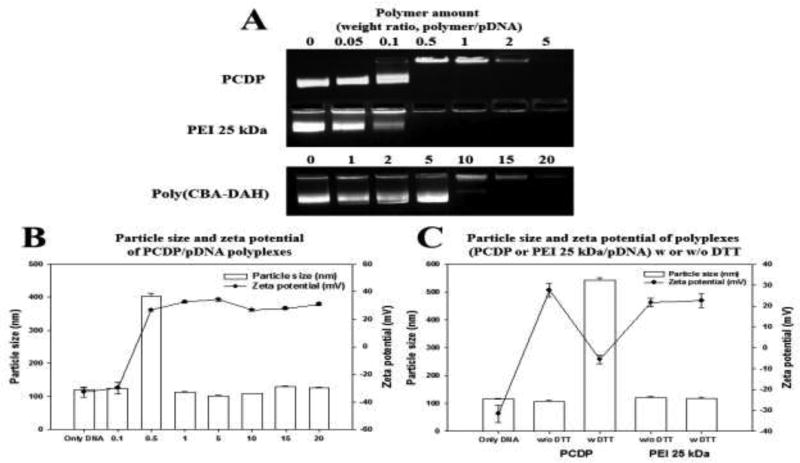
Naked pDNA prevent from entering the cell membrane due to their hydrophilicity and negative charge. The polycationic polymer, with its ability to form polyplexes with pDNA, provides a way to overcome this problem. The binding ability between pDNA and PCPD was evaluated by retardation of the pDNA (gWiz-Luc) in gel electrophoresis, displayed in Figure 2(A). PEI 25 kDa and poly(CBA-DAH) were used as comparison groups. The bend of pDNA, which formed the polyplexes with poly(CBA-DAH), displayed pDNA migration at weight ratio of 10 or 15. On the other hand, PCDP could bind pDNA thoroughly at a very low weight ratio of 0.5 and even displayed a characteristics similar to PEI 25 kDa. This gel retardation assay data indicated that PCDP had better DNA binding capacity than poly(CBA-DAH) due to increased amine groups from conjugation with PEI. This enhanced binding capacity can decrease the amount of polymer needed for successful delivery and easy application.
Figure 2.
(A) Agarose gel electrophoresis of PCDP, PEI 25 kDa, and poly(CBA-DAH) polyplexes with pDNA (gWiz-Luc). The number (0) means only naked pDNA and 0.05 to 5 (PCDP and PEI 25 kDa) and 1 to 20 (poly(CBA-DAH)) mean weight ratio based on pDNA (300 ng). (B) The particle size and zeta-potential of PCDP/pDNA polyplexes at various weight ratio ranging from 0.1 to 20 based on pDNA (gWiz-Luc, 4 µg). (C) Particle size and zeta-potential of PCDP/pDNA polyplexes at a weight ratio 10 with 5 mM DTT to confirm the dissociation of polyplexes. PEI 25 kDa/pDNA polyplexes (weight ratio 1) were used as a comparison group. Results are represented as mean ± SD. (n=3).
The positive surface charge and appropriate size (50 to several hundred nanometers) of polyplexes are important factors that affect the cellular uptake of the polyplexes (Y. Liu & Reineke, 2005; K. Nam et al., 2015; Zhang et al., 2015). PCDP/pDNA polyplexes were formed by electrostatic interaction between PCDP and pDNA (gWiz-Luc) at various weight ratios. The particle size and zeta potential results of PCDP/pDNA polyplexes are displayed in Figure 2(B). The particle sizes and zeta potentials of PCDP/pDNA were about 102 ~ 128 nm and 27 ~ 34 mV at weight ratios of 0.5 to 20, respectively. Poly(CBA-DAH)/pDNA had the particle size and zeta potentials of about 150 ~ 200 nm and 40 mV in our previously works (T. I. Kim et al., 2009; Ou et al., 2008). This decreased particle size of PCDP/pDNA polyplexes is considered to be due to the formation of more tightly polyplexes compared to poly(CBA-DAH)/pDNA polyplexes because of the increased presence of amine groups in PCDP. The negative value (−30 mV) of PCDP was measured at a weight ratio of 0.1, suggesting that the polyplexes are not formed at that ratio. At a weight ratio of 0.5, the zeta potential was dramatically increased to 27 mV. This result indicated that polyplexes can form from weight ratios above 0.5, which is also consistent with the gel retardation assay. However, the particle size was about 400 nm due to the unstable polyplexes formation and big cluster formation by self-aggregation (Zhang et al., 2015). The stable polyplexes of PCDP was formed at weight ratio of 1 and above.
The polycationic polymer which contains disulfide bonds can easily release pDNA after cleaved disulfide bond by reducing reagent (Doss et al., 2013; Oupicky & Li, 2014). To confirm the change in polyplexes through the cleaved disulfide bond of PCDP, DTT (5 mM) was used as an intracellular reducing agent like GSH. As shown in Figure 2(C), the particle size and zeta potential of PEI 25 kDa/pDNA polyplexes were not influenced by 5 mM DTT. Meanwhile, the particle size of PCPD/pDNA polyplexes with 5 mM DTT dramatically increased to 542 nm and zeta potential was negative charged (−6 mV). The cleavage of the disulfide bond of bio-reducible polymer by reducing agent increased the particle size due to the decreased the binding ability (Wen et al., 2011; Yu, Russ, & Wagner, 2009).
3.4. Cytotoxicity of PCPD and PCDP/pDNA polyplexes
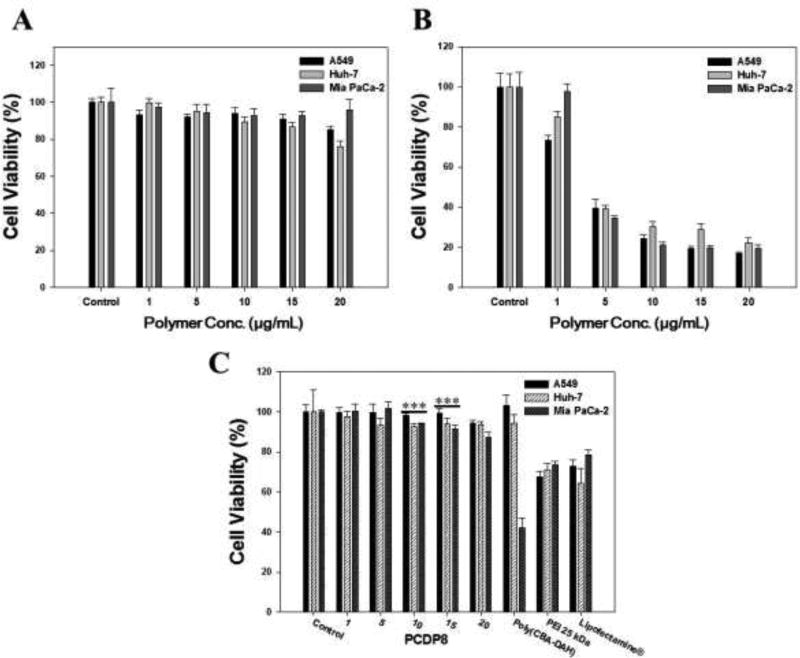
To confirm the cytotoxicity of polymer and polyplexes in A549, Huh-7, and Mia PaCa-2 cells, the MTT assay was performed. When the concentration of the treated PCPD was increased from 1 to 20 µg/mL, the cell viability was slightly decreased, as shown in Figure 3(A). However, the used maximum amount of PCDP to form the polyplexes with pDNA was 10 µg/mL and the cell viability was above 90% except in Huh-7 cells. As shown in Figure 3(B), the cell viability of PEI 25 kDa as a comparison group decreased significantly with increasing concentration up to 20 µg/mL. Generally, despite the increased transfection efficiency, PEI conjugated polycationic derivatives always had a problem with toxicity even PEI 600 Da or PEI 1.8 kDa due to the increased molecular weight from conjugation or polymerization. PCDP had no considerable cytotoxicity because PCDP consists of disulfide and amide bonds that can be degraded into non-toxic small molecules in cells. Many works have been published about biodegradable polymeric gene carriers that showed that biodegradability can reduce their cytotoxicity (T. I. Kim et al., 2009; C. Liu et al., 2012; Petersen, Merdan, Kunath, Fischer, & Kissel, 2002; Yu et al., 2009; Zhao, Roesler, & Kissel, 2011). As shown in Figure 3(C), the PCDP/pDNA polyplexes were also observed to have no considerable cytotoxicity at all polyplexes formation ratios (w/w, 1 to 20) in all cell lines. However, the comparison groups such as PEI 25 kDa and Lipofectamine® polyplexes showed below 80% cell viability in all cell lines. In the case of poly(CBA-DAH) polyplexes, there was no toxicity in A549 and Huh-7 cells, whereas the cell viability decreased to 42% in Mai PaCa-2 cell. This result indicates that the poly(CBA-DAH) is seriously toxic to certain cells. The results of MTT assay indicate that PCDP had no cytotoxicity and that it should be safer than comparison groups when it is used to deliver the pDNA.
Figure 3.
Cell viability of polymer (A) only PCDP and (B) only PEI 25 kDa and various polyplexes (C) in A549, Huh-7, and Mia PaCa-2 cells. The numbers in the bottom refer to the mean concentration of polymer ranging from 1 to 20 µg/mL (A and B) and weight ratios of PCDP/pDNA polyplexes based on pDNA (gWiz-Luc, 500 ng) (C). Poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® were formed the polyplexes with pDNA at weight ratio 1:40, 1:1, and 1:2, respectively. Results are represented as mean ± SD. (n=3). ***P <0.001 versus PEI 25 kDa.
3.5. Cellular uptakes of PCDP/pDNA polyplexes
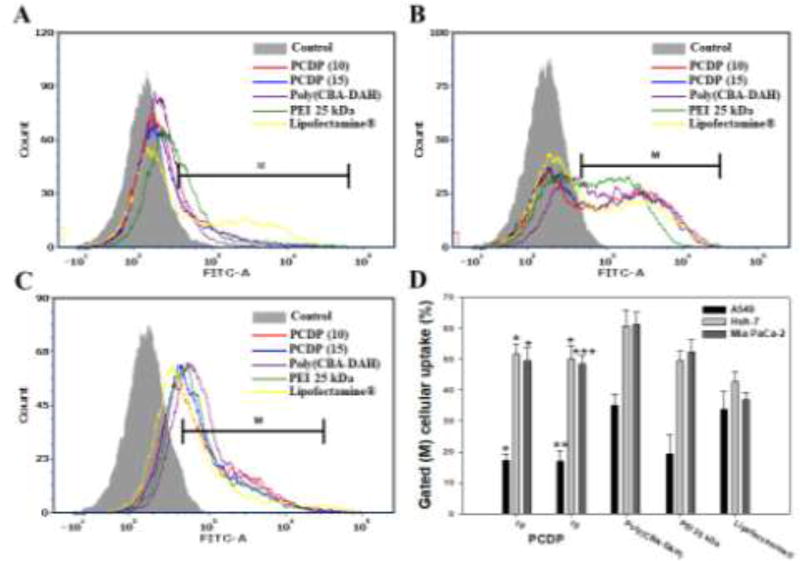
The cellular uptakes of PCDP/pDNA polyplexes were investigated at various weight ratios (1 to 20) by FACscan analyzer in various cancer cell lines. The polyplexes were formed between polymers and pDNA which was stained by YOYO-1 dye according to the manufacturer’s protocol for green fluorescence measurements. Poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® were also used as comparison groups and formed polyplexes at optimized or recommended weight ratio of 40, 1, and 2 based on pDNA (1 µg), respectively. The cellular uptake efficiency of PCPD polyplexes showed low efficiency at weight ratio of 1 and 5, but showed almost similar efficiency at weight ratios of 10 and above in all cell lines (data not shown). To compare with control groups, the cellular uptake efficiency of PCDP polyplexes at weight ratio of 10 and 15 are displayed in Figure 4.
Figure 4.
Cellular uptake of polyplexes in (A) A549, (B) Huh-7, and (C) Mia PaCa-2 cells. The YOYO-1 stained pDNA were formed the polyplexes with PCDP, poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® at weight ratio based on pDNA (1 µg), respectively. (D) The cellular uptake % of quantification of cell internalization measured by FACs. Results are represented as mean ± SD. (n=3). *<0.05, **P < 0.01, ***P <0.001 versus PEI 25 kDa.
As shown in Figure 4, PCDP polyplexes showed similar cellular uptake and quantified cellular uptake (%) with PEI 25 kDa polyplexes in all cell lines and showed better results than Lipofectamine® except that of A549 cells. All tested polymers can form polyplexes with nano-sized particle size and positive surface charge. The particle sizes and zeta potentials were 102 ~ 128 nm and 27 ~ 34 mV (PCDP), 120 nm and 22 mV (PEI 25 kDa), <200 nm and 40 mV (poly(CBA-DAH)) (T. I. Kim et al., 2009; Ou et al., 2008), and 150 nm and 38 mV (Lipofectamine®) (Lee et al., 2008), respectively. In the case of poly(CBA-DAH) polyplexes, showed highest cellular uptake efficiency in all cell lines. This result was considered to be due to the highest positive charge of poly(CBA-DAH), as well as other factors including shape or surface hydrophobicity. The cellular uptake was affected by not only particle size and positive surface charge but also materials, surface hydrophobicity and shape (Frohlich, 2012; J. P. Nam et al., 2015).
3.6. In-vitro transfection experiment
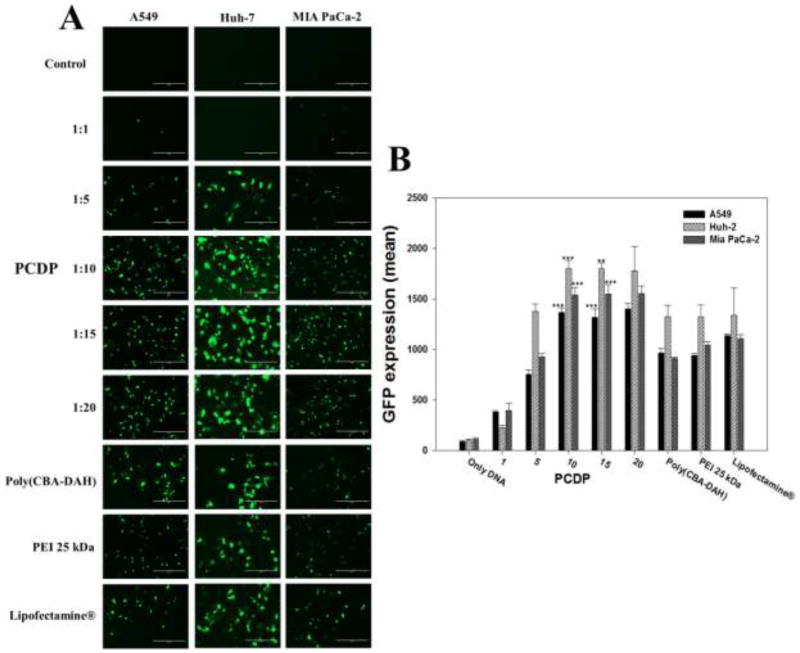
To evaluate the gene transfection efficiency of PCDP polyplexes, both reporter genes gWiz-Luc and gWiz-GFP were used and formed polyplexes with PCDP. The comparison groups including poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® were also treated in various cancer cell lines. The direct visualization of GFP expression of polymer/pDNA (gWiz-GFP) polyplexes are displayed in Figure 5. The GFP gene expression of PCDP polyplexes were increased with increasing polyplexes formation ratios ranging from 1 to 10 (w/w ratio) but showed almost similar results above weight ratio of 10 in all treated cell lines. This result indicates that the optimal ratio are at weight ratio of 10 or 15. Therefore, the following experiments such as luciferase and VEGF silencing assay of PCDP polyplexes were measured at the optimal weight ratios of 10 and 15. The comparison groups including poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® showed similar GFP gene expression in all cell lines. In the case of PCDP above 10 (w/w ratio), the visualized GFP gene expression and the mean fluorescence intensity (MFI) were higher than other comparison groups. As described above, PCDP has disulfide bonds which can be cleaved by reducing agents as GSH, allowing pDNA to be easily released from polyplexes into the cytoplasm with no considerable cytotoxicity. Therefore, PCDP polyplexes had highest GFP gene expression efficiency. In addition, poly(CBA-DAH) polyplexes had more cellular uptake than PCDP polyplexes, but PCDP polyplexes showed the increased GFP gene expression efficiency. It may be considered that it can easily escape from the endosome due to increased buffering capacity by PEI conjugation. The MFI of PCDP polyplexes in cell lines followed the rank Huh-7>Mia PaCa-2>A549 cells.
Figure 5.
(A) The monitored by fluorescent microscopy GFP expression of polyplexes against A549, Huh-7, and Mia PaCa-2 cells for 48 h. (B) The quantified and normalized GFP expression (mean). The scale bar (A) means 200 µm and the numbers (B) in the bottom refer to the mean the weight ratios based on pDNA (gWiz-GFP, 500 ng). Results are represented as mean ± SD (n = 3). **P < 0.01, ***P <0.001 versus PEI 25 kDa.
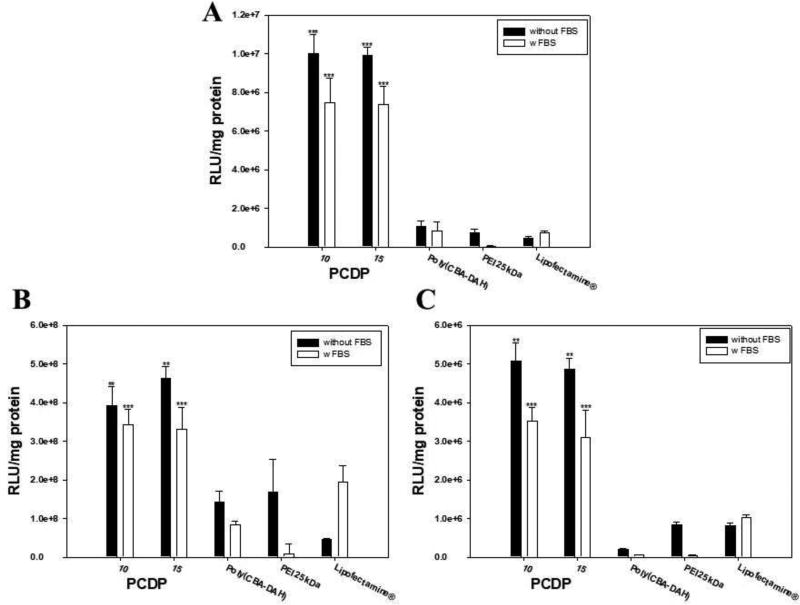
Figure 6 shows the data of the transfection efficiency using gWiz-Luc. In addition, to mimic in vivo conditions, the transfection efficiency was also evaluated in the presence of FBS (30%). The related light units (RLU) of PCDP at both weight ratio 10 and 15 were higher than other comparison groups in all treated cell lines. These results were consistent with MFI results of the GFP expression assay. The RLU of PCDP, poly(CBA-DAH), and PEI 25 kDa polyplexes was decreased in mimicked in vivo conditions. Interestingly, Lipofectamine® showed increased transfection efficiency at the same conditions, where this phenomenon was also shown in our previously work (K. Nam et al., 2015). However, the transfection efficiency of PCDP polyplexes was still higher than other comparison groups, even Lipofectamine® polyplexes, which had having increased transfection efficiency in mimicked in vivo conditions. Especially, PCDP polyplexes showed 10.2 times, 1.7 times, and 6.1 times higher gene transfection efficiency than Lipofectamine® polyplexes in A549, Huh-7, and Mia PaCa-2 cells, respectively. The RLC of PCDP polyplexes in cell lines with or without FBS followed the rank Huh-7>A549>Mia PaCa-2 cells.
Figure 6.
Luciferase expression of polymer/pDNA (gWiz-Luc, 500 ng) polyplexes in (A) A549, (B) Huh-7, and (C) Mia PaCa-2 cells for 48 h. Results are represented as mean ± SD. (n=3). Black bars mean (−) FBS and white bars mean (+) FBS. **P < 0.01, ***P <0.001 versus PEI 25 kDa.
3.7. VEGF silencing and cancer cell growth inhibition efficiency of PCDP/pshVEGF
The VEGF siRNA expressing plasmid (pshVEGF) was constructed to inhibit VEGF expression of cancer cells and to express VEGF siRNA longer than directly delivering siRNA via methods mentioned above from our previous work (H. A. Kim et al., 2014). VEGF is well known a signal protein produced by cells that stimulates vasculogenesis and angiogenesis. The cancer cells can express VEGF, leading to the growth and metastasis of tumor via angiogenesis. Therefore, disabling VEGF receptor function and inhibiting VEGF expression has been used as a strategy to inhibit tumor growth and metastasis (H. A. Kim et al., 2014; Yang et al., 2011; Zibara et al., 2015).
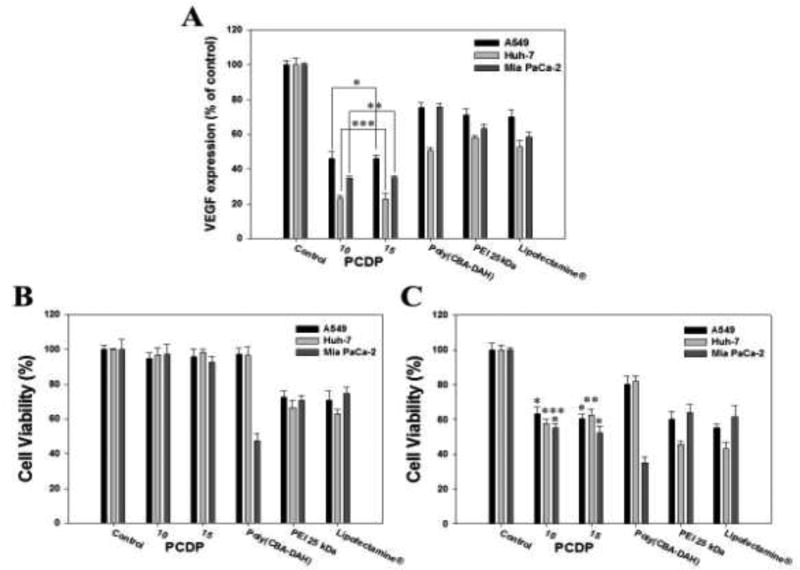
The VEGF gene expressions (% of control) in relation with gene silencing efficiency were measured by human VEGF ELISA in Figure 7(A). The cells were transfected by polymers/pshVEGF polyplexes. The VEGF expressions of PCDP polyplexes at weight ratio of 10 and 15 were dramatically decreased and showed almost similar results between weight ratio of 10 and 15. In addition, the PCDP polyplexes showed lower VEGF expression than poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® polyplexes in all cell lines and these results were consistent with transfection efficiency assay using gWiz-Luc and gWiz-GFP. Compared to non-treated control cells, gene silencing efficiency of PCDP polyplexes showed 54%, 77%, and 66% (w/w; 10) and 55%, 78%, and 65% (w/w; 15) reduction of VEGF levels in A549, Huh-7, and Mia PaCa-2 cells, respectively.
Figure 7.
Down regulation of VEGF expression using pshVEGF delivery into the cancer cells for 48 h by polymers. The VEGF expression were measured by human VEGF ELISA kit. (A) Quantification of down regulated VEGF expression shown by relative VEGF expression %. Relative VEGF expression (%) = Amount of VEGF (Treated)/Amonut of VEGF (Control) × 100. Cell growth inhibition of polymers with gWiz-Luc (B) or shVEGF (C) polyplexes were measured by MTT assay. Results are represented as mean ± SD. (n=3). *P<0.05, **P < 0.01, ***P <0.001 versus PEI 25 kDa.
The cells growth inhibition of PCPD/pshVEGF polyplexes was investigated at the same experimental condition as in ELISA. In addition, to confirm whether or not the inhibition of cell growth was due to VEGF gene silencing or polymer toxicity, the polyplexes using gWiz-Luc was treated into the cells with the same conditions and measured by MTT assay. The cell viabilities of PCDP polyplexes were decreased from 95% (formed with gWiz-Luc) to 60% (formed with pshVEGF) as shown in Figure 7(B and C). This result indicates that the VEGF gene silencing by PCDP/pshVEGF polyplexes indirectly inhibits the cell proliferation and growth rates. The comparison groups, which used pshVEGF, also showed decreased cell viability, even lower than that of PCDP polyplexes. However, the decrease in cell viability is not only from the VEGF siRNA silencing effect but also from the cytotoxicity of the polymers (Figure 7(B)). The reduced cell viability rates associated with both results (gWiz and pshVEGF) were 31%, 39%, and 42% (PCDP, w/w; 10), 35%, 36%, and 40% (PCDP, w/w; 15), 17%, 14%, and 12% (poly(CBA-DAH)), 12%, 21%, and 6% (PEI 25 kDa), and 15%, 20%, and 13% (Lipofectamine®) in A549, Huh-7, and Mia PaCa-2 cells, respectively. The PCDP polyplexes with weight ratio of 10 or 15 had similar results in the all experiments. Therefore, the optimal weight ratio of PCDP is 10.
4. Conclusion
As mentioned above, PEI has many nitrogen atoms including primary, secondary, and tertiary amine groups, which can increase binding affinity with pDNA, buffering capacity and provide positive charge to polyplexes, where theses amine groups increase cellular uptake and endosomal escape, leading to increase the transfection efficiency.
In this study, we synthesized the PEI-conjugated poly(CBA-DAH) (PCDP) through disulfide bonds to decrease the amount needed for formation of polyplexes with pDNA and to increase the transfection efficiency. The synthesized PCDP showed great pDNA binding ability in very low weight ratios, similar to PEI 25 kDa. As per results of gene transfection efficiency assay using gWiz-Luc and gWiz-GFP, PCDP polyplexes had higher transfection efficiency than comparison groups including poly(CBA-DAH), PEI 25 kDa, and Lipofectamine® polyplexes in all tested cancer cell lines. To confirm the tumor cell growth inhibition, pshVEGF (VEGF siRNA expressing plasmid) was used and transfected by PCDP into A549, Huh-7, and Mia PaCa-2 cells. The results showed that the VEGF expression of PCDP polyplexes was dramatically decreased than comparison groups, leading to a decrease in cell viability. Therefore, the synthesized PCDP may be a strong candidate as a pDNA delivery carrier for gene therapy.
Acknowledgments
This work was supported by the NIH Grant CA177932 (S.W. Kim) and Basic Sciency Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education 2015R1A6A3A03017256 (J-P. Nam).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interest.
References
- Al-Dosari MS, Gao X. Nonviral gene delivery: principle, limitations, and recent progress. AAPS J. 2009;11(4):671–681. doi: 10.1208/s12248-009-9143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello Roufai M, Midoux P. Histidylated polylysine as DNA vector: elevation of the imidazole protonation and reduced cellular uptake without change in the polyfection efficiency of serum stabilized negative polyplexes. Bioconjug Chem. 2001;12(1):92–99. doi: 10.1021/bc0000738. [DOI] [PubMed] [Google Scholar]
- Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The possible "proton sponge " effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol Ther. 2013;21(1):149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouard D, Alazard-Dany D, Cosset FL. Viral vectors: from virology to transgene expression. Br J Pharmacol. 2009;157(2):153–165. doi: 10.1038/bjp.2008.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J, Drevin H, Axen R. Protein thiolation and reversible protein-protein conjugation. N-Succinimidyl 3-(2-pyridyldithio)propionate, a new heterobifunctional reagent. Biochem J. 1978;173(3):723–737. doi: 10.1042/bj1730723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EM BO Rep. 2006;7(3):271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss CG, Debottam S, Debajyoti C. Glutathione-responsive nano-transporter-mediated siRNA delivery: silencing the mRNA expression of Ras. Protoplasma. 2013;250(3):787–792. doi: 10.1007/s00709-012-0451-1. [DOI] [PubMed] [Google Scholar]
- Eroglu E, Tiwari PM, Waffo AB, Miller ME, Vig K, Dennis VA, Singh SR. A nonviral pHEMA+chitosan nanosphere-mediated high-efficiency gene delivery system. Int J Nanomedicine. 2013;8:1403–1415. doi: 10.2147/IJN.S43168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Bieber T, Li Y, Elsasser HP, Kissel T. A novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicity. Pharm Res. 1999;16(8):1273–1279. doi: 10.1023/a:1014861900478. [DOI] [PubMed] [Google Scholar]
- Frohlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomedicine. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan SS, Rechnitz GA. New liquid membrane electrode for the determination of ephedrine, epinephrine, and norepinephrine. Anal Chem. 1986;58(6):1052–1054. doi: 10.1021/ac00297a015. [DOI] [PubMed] [Google Scholar]
- Hong R, Han G, Fernandez JM, Kim BJ, Forbes NS, Rotello VM. Glutathione-mediated delivery and release using monolayer protected nanoparticle carriers. J Am Chem Soc. 2006;128(4):1078–1079. doi: 10.1021/ja056726i. [DOI] [PubMed] [Google Scholar]
- Katz MG, Fargnoli AS, Williams RD, Bridges CR. Gene therapy delivery systems for enhancing viral and nonviral vectors for cardiac diseases: current concepts and future applications. Hum Gene Ther. 2013;24(11):914–927. doi: 10.1089/hum.2013.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HA, Nam K, Kim SW. Tumor targeting RGD conjugated bio-reducible polymer for VEGF siRNA expressing plasmid delivery. Biomaterials. 2014;35(26):7543–7552. doi: 10.1016/j.biomaterials.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TI, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30(4):658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layek B, Singh J. Amino acid grafted chitosan for high performance gene delivery: comparison of amino acid hydrophobicity on vector and polyplex characteristics. Biomacromolecules. 2013;14(2):485–494. doi: 10.1021/bm301720g. [DOI] [PubMed] [Google Scholar]
- Lee JH, Ahn HH, Kim KS, Lee JY, Kim MS, Lee B, Lee HB. Polyethyleneimine-mediated gene delivery into rat pheochromocytoma PC-12 cells. J Tissue Eng Regen Med. 2008;2(5):288–295. doi: 10.1002/term.94. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhu Q, Wu W, Xu X, Wang X, Gao S, Liu K. Degradable copolymer based on amphiphilic N-octyl-N-quatenary chitosan and low-molecular weight polyethylenimine for gene delivery. Int J Nanomedicine. 2012;7:5339–5350. doi: 10.2147/IJN.S36179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. J Am Chem Soc. 2005;127(9):3004–3015. doi: 10.1021/ja0436446. [DOI] [PubMed] [Google Scholar]
- Luu QP, Shin JY, Kim YK, Islam MA, Kang SK, Cho MH, Cho CS. High gene transfer by the osmotic polysorbitol-mediated transporter through the selective caveolae endocytic pathway. Mol Pharm. 2012;9(8):2206–2218. doi: 10.1021/mp300072r. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nam HY, Nam K, Lee M, Kim SW, Bull DA. Dendrimer type bio-reducible polymer for efficient gene delivery. J Control Release. 2012;160(3):592–600. doi: 10.1016/j.jconrel.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Nam JP, Nam K, Nah JW, Kim SW. Evaluation of Histidylated Arginine-Grafted Bioreducible Polymer To Enhance Transfection Efficiency for Use as a Gene Carrier. Mol Pharm. 2015;12(7):2352–2364. doi: 10.1021/acs.molpharmaceut.5b00013. [DOI] [PubMed] [Google Scholar]
- Nam JP, Park JK, Son DH, Kim TH, Park SJ, Park SC, Nah JW. Evaluation of polyethylene glycol-conjugated novel polymeric anti-tumor drug for cancer therapy. Colloids Surf B Biointerfaces. 2014;120:168–175. doi: 10.1016/j.colsurfb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- Nam K, Jung S, Nam JP, Kim SW. Poly(ethylenimine) conjugated bioreducible dendrimer for efficient gene delivery. J Control Release. 2015;220(Pt A):447–455. doi: 10.1016/j.jconrel.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res. 2012;1:27. doi: 10.4103/2277-9175.98152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9(24):1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- Ou M, Wang XL, Xu R, Chang CW, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjug Chem. 2008;19(3):626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oupicky D, Li J. Bioreducible polycations in nucleic acid delivery: past, present, and future trends. Macromol Biosci. 2014;14(7):908–922. doi: 10.1002/mabi.201400061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack DW, Putnam D, Langer R. Design of imidazole-containing endosomolytic biopolymers for gene delivery. Biotechnol Bioeng. 2000;67(2):217–223. [PubMed] [Google Scholar]
- Petersen H, Merdan T, Kunath K, Fischer D, Kissel T. Poly(ethylenimine-co-L-lactamide-co-succinamide): a biodegradable polyethylenimine derivative with an advantageous pH-dependent hydrolytic degradation for gene delivery. Bioconjug Chem. 2002;13(4):812–821. doi: 10.1021/bc0255135. [DOI] [PubMed] [Google Scholar]
- Pires LR, Oliveira H, Barrias CC, Sampaio P, Pereira AJ, Maiato H, Pego AP. Imidazole-grafted chitosan-mediated gene delivery: in vitro study on transfection, intracellular trafficking and degradation. Nanomedicine (Lond) 2011;6(9):1499–1512. doi: 10.2217/nnm.11.51. [DOI] [PubMed] [Google Scholar]
- Stone D. Novel viral vector systems for gene therapy. Viruses. 2010;2(4):1002–1007. doi: 10.3390/v2041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Gao L, Meng LY, Xie JM, Xiong JW, Luo Y. A Neutralized Noncharged Polyethylenimine-Based System for Efficient Delivery of siRNA into Heart without Toxicity. ACS Appl Mater Interfaces. 2016;8(49):33529–33538. doi: 10.1021/acsami.6b13295. [DOI] [PubMed] [Google Scholar]
- Wen HY, Dong HQ, Xie WJ, Li YY, Wang K, Pauletti GM, Shi DL. Rapidly disassembling nanomicelles with disulfide-linked PEG shells for glutathione-mediated intracellular drug delivery. Chem Commun (Camb) 2011;47(12):3550–3552. doi: 10.1039/c0cc04983b. [DOI] [PubMed] [Google Scholar]
- Xu Q, Wang CH, Pack DW. Polymeric carriers for gene delivery: chitosan and poly(amidoamine) dendrimers. Curr Pharm Des. 2010;16(21):2350–2368. doi: 10.2174/138161210791920469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Liu X, Zhang D, Yu W, Lv G, Xie H, Ma X. Chitosan/VEGF-sIRNA nanoparticle for gene silencing. J Control Release. 2011;152(Suppl 1):e160–161. doi: 10.1016/j.jconrel.2011.08.062. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xu Z, Chen S, Gao Y, Gu W, Chen L, Li Y. Histidylated cationic polyorganophosphazene/DNA self-assembled nanoparticles for gene delivery. Int J Pharm. 2008;353(1–2):277–282. doi: 10.1016/j.ijpharm.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Yu H, Russ V, Wagner E. Influence of the molecular weight of bioreducible oligoethylenimine conjugates on the polyplex transfection properties. AAPS J. 2009;11(3):445–455. doi: 10.1208/s12248-009-9122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Duan Y, Wang D, Bian F. Preparation of arginine modified PEI-conjugated chitosan copolymer for DNA delivery. Carbohydr Polym. 2015;122:53–59. doi: 10.1016/j.carbpol.2014.12.054. [DOI] [PubMed] [Google Scholar]
- Zhao N, Roesler S, Kissel T. Synthesis of a new potential biodegradable disulfide containing poly(ethylene imine)-poly(ethylene glycol) copolymer cross-linked with click cluster for gene delivery. Int J Pharm. 2011;411(1–2):197–205. doi: 10.1016/j.ijpharm.2011.03.038. [DOI] [PubMed] [Google Scholar]
- Zibara K, Awada Z, Dib L, El-Saghir J, Al-Ghadban S, Ibrik A, El-Sabban M. Anti-angiogenesis therapy and gap junction inhibition reduce MDA-MB-231 breast cancer cell invasion and metastasis in vitro and in vivo. Sci Rep. 2015;5:12598. doi: 10.1038/srep12598. [DOI] [PMC free article] [PubMed] [Google Scholar]