Abstract
Plasmodium falciparum dihydrofolate reductase (pf-DHFR) is one of the several targets in the treatment of malaria. Double and quadruple mutations at residues 51, 59, 108, and 164 of pf-DHFR have been linked to antifolate resistance. Several efforts are underway to overcome this drug resistance and to produce potential inhibitors. In this regard, the quantitative structure-activity relationship (QSAR) and docking studies were performed for previously reported 4-anilinoquinoline and 1,3,5-triazines based molecular hybrids. The generated model showed good correlation coefficients (R2 = 0.70) and test set prediction coefficient (R2 = 0.74). These outcomes showed the good predictive competence of the established QSAR model. Based on these results we docked into active site of pf-DHFR protein with the most active (4) and the less active (5) compounds. The docking results revealed that these molecules interact specifically with SER108 and ILE164 in the pf-DHFR binding pocket as that of best active compound but also showed additional interactions with LEU40 and GLY44.
Keywords: Theoretical chemistry, Pharmaceutical chemistry, Molecular docking, Antimalarial activity, pf-DHFR, QSAR, 4-Anilinoquinoline-triazine
1. Introduction
Malaria is an infectious disease caused by parasites of the genus Plasmodium [1]. Five species of malaria parasites are known, namely, P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi [2]. Approximately 90 % of deaths (generally of children in Africa) related to malaria infections are caused by Plasmodium falciparum [3, 4]. In 2016, 216 million of human beings are counted by the World Health Organization (WHO) over 91 countries of the world, which are affected by malaria, with an increase of 5 million cases compared to previous years [5].
Despite of the great efforts devoted to discover an effective antimalarial drugs, these efforts suffer from many obstacles, including drug resistance issues [6, 7]. To overcome this problem, the concept of hybrid molecules has been introduced as one of the most used solutions, in which two or more pharmacophores are linked together and act by inhibiting simultaneously two conventional targets [8]. In this regard, the 1,3,5-triazine derivatives such as cycloguanil, chlorcycloguanil and WR99210are already approved as effective dihydrofolatereductase (DHFR) inhibitors, which selectively inhibit biochemical processes that are vital for parasite growth [9]. In addition, quinoline nucleus has attracted much interest of medicinal chemists, as an imperative pharmacophore accountable for imparting antimalarial action [10, 11]. Furthermore, to become a drug a novel synthetic molecule must take a very long journey. As a consequence, the pharmaceutical industry is moving towards new research methods, involving predicting the activities of molecules before they are even synthesized. The use of molecular modeling techniques such as QSAR and molecular docking has produced very impressive results in last year's [12, 13]. In the purpose to pursue our previous works [14, 15]. In this paper, we have performed the molecular modeling of 4-anilinoquinoline-triazines (Fig. 1) as a potential antimalarial compounds by using QSAR and docking studies [16].
Fig. 1.
The structure of the 4-anilinoquinoline-triazine derivatives.
Quantitative Structure Activity Relationships (QSAR) is very important tool in drug discovery [17]. The multidimensional molecular descriptors (1D, 2D, 3D) have been calculated to identify regions in space which are correlated to the biological activities (Table 1) [18]. QSAR models could be generated by using statistical methods. In this study, we have used a training set of 37 4-anilinoquinoline-triazines derivatives to build the QSAR model [19]. For this purpose, we used multiple linear regression analysis (MLR)) and artificial neural networks (ANN). The predictive ability of the established model has been tested by several validation techniques such as: internal and external validations as well as Y-randomization methods. Plasmodium falciparum dihydrofolate reductase (pf-DHFR) is the most important targets for antimalarial drug discovery. In addition, we have performed the docking molecular of two compounds, 4 and 5, with Pf-DHFR in its two forms; the wild type and the quadruple mutant [20]. Also we investigated the interaction of these hybrids with the binding site of Pf-DHFR protein structures, in order to gain structural insight for improved antimalarial activity.
Table 1.
Studied compounds and their observed activities logIC50.
| Compounds | R1 | R2 | IC50(obs) | LogIC50(obs) |
|---|---|---|---|---|
| 1 | 3-fluoroanilinyl | 4-methylpiperazin-1-yl | 22.80 | 1.358 |
| 2 | 3-fluoroanilinyl | 4-ethylpiperazin-1-yl | 2.47 | 0.393 |
| 3 | 3-fluoroanilinyl | 2-morpholinoethan-1-aminyl | 40.02 | 1.602 |
| 4 | 3-fluoroanilinyl | 3-morpholinopropan-1-aminyl | 1.36 | 0.134 |
| 5 | 3-fluoroanilinyl | piperidin-1-yl | 128.74 | 2.110 |
| 6 | 4-fluoroanilinyl | 4-methylpiperazin-1-yl | 8.97 | 0.953 |
| 7 | 4-fluoroanilinyl | 4-ethylpiperazin-1-yl | 11.11 | 1.046 |
| 8 | 4-fluoroanilinyl | 2-morpholinoethan-1-aminyl | 9.81 | 0.992 |
| 9 | 4-fluoroanilinyl | 3-morpholinopropan-1-aminyl | 15.82 | 1.199 |
| 10 | 4-fluoroanilinyl | piperidin-1-yl | 41.75 | 1.621 |
| 11 | 3-chloroanilinyl | 4-methylpiperazin-1-yl | 2.41 | 0.382 |
| 12 | 3-chloroanilinyl | 4-ethylpiperazin-1-yl | 2.11 | 0.324 |
| 13 | 3-chloroanilinyl | 2-morpholinoethan-1-aminyl | 30.60 | 1.486 |
| 14 | 3-chloroanilinyl | 3-morpholinopropan-1-aminyl | 3.82 | 0.582 |
| 15 | 4-chloroanilinyl | 4-methylpiperazin-1-yl | 7.80 | 0.892 |
| 16 | 4-chloroanilinyl | 4-ethylpiperazin-1-yl | 4.14 | 0.617 |
| 17 | 4-chloroanilinyl | 3-morpholinopropan-1-aminyl | 4.63 | 0.666 |
| 18 | 3-methoxyanilinyl | 4-methylpiperazin-1-yl | 4.26 | 0.629 |
| 19 | 3-methoxyanilinyl | 4-ethylpiperazin-1-yl | 8.90 | 0.949 |
| 20 | 3-methoxyanilinyl | 3-morpholinopropan-1-aminyl | 2.98 | 0.474 |
| 21 | 4-methoxyanilinyl | 4-ethylpiperazin-1-yl | 4.26 | 0.629 |
| 22 | 4-methoxyanilinyl | 3-morpholinopropan-1-aminyl | 3.81 | 0.581 |
| 23 | 3,4-dimethoxyanilinyl | 4-methylpiperazin-1-yl | 5.04 | 0.702 |
| 24 | 3,4-dimethoxyanilinyl | 4-ethylpiperazin-1-yl | 2.58 | 0.412 |
| 25 | 3,4-dimethoxyanilinyl | 3-morpholinopropan-1-aminyl | 4.58 | 0.661 |
| 26 | 1,2,3,4-tetrahydroquinolinyl | 4-methylpiperazin-1-yl | 12.64 | 1.102 |
| 27 | 1,2,3,4-tetrahydroquinolinyl | 4-ethylpiperazin-1-yl | 20.84 | 1.319 |
| 28 | 1,2,3,4-tetrahydroquinolinyl | 2-morpholinoethan-1-aminyl | 17.35 | 1.239 |
| 29 | 1,2,3,4-tetrahydroquinolinyl | 3-morpholinopropan-1-aminyl | 14.70 | 1.167 |
| 30 | 1,2,3,4-tetrahydroisoquinolin-2(1H)-yl | 4-methylpiperazin-1-yl | 28.12 | 1.449 |
| 31 | 1,2,3,4-tetrahydroisoquinolin-2(1H)-yl | 4-ethylpiperazin-1-yl | 12.12 | 1.084 |
| 32 | 1,2,3,4-tetrahydroisoquinolin-2(1H)-yl | 2-morpholinoethan-1-aminyl | 45.21 | 1.655 |
| 33 | 1,2,3,4-tetrahydroisoquinolin-2(1H)-yl | 3-morpholinopropan-1-aminyl | 20.32 | 1.308 |
| 34 | morpholinyl | 4-methylpiperazin-1-yl | 3.22 | 0.508 |
| 35 | morpholinyl | 4-ethylpiperazin-1-yl | 2.16 | 0.334 |
| 36 | morpholinyl | 2-morpholinoethan-1-aminyl | 5.87 | 0.769 |
| 37 | morpholinyl | 3-morpholinopropan-1-aminyl | 2.76 | 0.441 |
2. Materials and methods
2.1. Studied molecules
To perform the molecular modeling we have taken the experimental antimalarial activities data of 37 hybrids molecules described previously [16]. Thus, the observed activities (IC50) are converted into logarithm scale logIC50 and they are presented in Table 1.
2.2. Molecular descriptors calculation
In order to build a reliable QSAR model, a total of 14 descriptors including lipophilic, geometrical, physicochemical, and steric. Those are calculated with the MM2 method using ACD/ChemSketch [21] and ChemBioOffice softwares [22]. Meanwhile, the geometry of the studied compounds was optimized using DFT/B3LYP(6-31G) method [23, 24]. The electronic descriptors were calculated by Gaussian 03 quantum chemistry software [25]. All descriptors used in this work are presented in Table 2.
Table 2.
List of the calculate descriptors.
| Type of descriptors | Electronic | lipophilic | Geometrical | physicochemical | Steric |
|---|---|---|---|---|---|
| Name of the descriptors | HOMO energy (EHOMO) LUMO energy (ELUMO) Dipole moment (Dp) total energy (E) |
lipophilic (Octanol-water partition coefficient) (LogP) VDW energy (EVDW) |
Torsion energy (T) stretch-bend energy (SB) |
Critical pressure (CP) Critical volume (CV) |
Density (D) Refractive Index (R) Surface Tension (ST) parchor (P) |
2.3. Statistical methods
In the aim to build QSAR model, we have chosen a set of 37 compounds from previously reported work, whose shown important antimalarial activity [16]. The complete set was randomly divided into two subsets a training set (29 compounds) to build the model and a test set (8 compounds) to evaluate the reliability of the established model. Various statistical methods were used to build the QSAR model, viz: Multiple Linear Regression (MLR) [26], and Artificial Neural Networks (ANN) [27]. Indeed, the MLR with descendent selection of variables was used to study the relation between one dependent variable (antimalarial activity) and several independent variables (calculated molecular descriptors). Furthermore, the MLR was used to select the descriptors that will serve as the input parameters for ANN. Hence, ANN could be considered as suitable tools that have powerful mechanism to capture patterns in data, which has been widely used to model nonlinear system [28]. In addition, the high values of the correlation coefficient indicate how the equations fit the data. On other hand, validation a strategy has been recognized to inquire into the applicability of the QSAR models on a new data. For this reason, we have used Cross-Validation with “leave-one-out” procedure, in order to explore the reliability of the proposed models. Cross validation was used in which a number of models were developed with one sample ignored each time. The model was evaluated by measuring its accuracy in predicting the responses of the remaining data (the ones that have not been used in the development of the model) [29, 30].
According to Golbraikh and Tropsha study on validation methods, cross-validation is necessary but not sufficient to ensure the predictive capability of the proposed QSAR model. In this study, we have validated the QSAR model by both MLR and ANN methods based on training set. The external validation should be in perfect agreement with the criteria of Golbraikh and Tropsha [31]. Finally, Y-randomization test has been used to exclude the possibility of random correlation between descriptors and its corresponding bioactivities in the obtained model. This test consists to mix randomly many properties/experimental activities for the new learning series using the same descriptors. For an acceptable QSAR model, the average correlation coefficient (Rr) of randomized models should be less than the correlation coefficient (R) of nonrandomized model [32].
2.4. Molecular docking modeling
A molecular modeling study was conducted in order to gain insight into the key structural requirements of a geometrical model and to analyze the interactions of the hybrid systems with the active sites of the protein pf-DHFR [14, 20] of both wild (coded as 1J3I.pdb) and quadruple mutant types (coded as 1J3K.pdb), which are obtained from the Protein Data Bank RCSB [33]. In this study, we have performed the molecular docking of two compounds with pf-DHFR protein. We have chosen the highest active compound (compound number 4) and the lowest active compound (compound number 5) of the studied series. First, we have removed all water molecules from the receptor, the ligands and non-protein parts by using the Discovery Studio software [34]. The AutoDock 4.2, has been used to analyze the interactions between the ligand and the protein [35]. The 3D grid was created by the AUTOGRID algorithm [36]to evaluate the interacting energy between ligands and wild-type protein. The grid maps were constructed using 60, 60 and 60 pointing in x, y and z directions, with grid point spacing of 0.375° A. The center grid box is about (30.323Å, 5.116Å and 58.385Å) by the ligand location in the complex. However, the center grid box of the quadruple mutant type protein is about (29.987Å, 5.56 Å and 57.424 Å). Discovery Studio software was used for the 2D and 3D visualizations of the established interactions [34].
3. Results and discussion
To perform this study, we have divided the 37 studied compounds randomly into training sets and test sets which containing 29 and 8 compounds respectively. The values of the selected descriptors and the predicted values of antimalarial activity of the training set which obtained by using MLR, ANN and CV methods are presented in Table 3.
Table 3.
The values of selected descriptors and observed/predicted activity (logIC50).
| N | E | S–B | T | EVDW | LogP | ST | LogIC50 (Obs) | MLR | ANN | Cv(LOO) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | -2213.007 | 0.520 | -15.547 | 42.004 | 7.347 | 72.100 | 0.393 | 0.7412 | 0.724 | 0.739 | |
| 3 | -2288.181 | 0.532 | -12.073 | 43.096 | 6.065 | 77.900 | 1.602 | 1.1958 | 1.311 | 1.299 | |
| 4 | -2327.486 | 0.556 | -12.039 | 43.729 | 6.170 | 75.700 | 0.134 | 0.8582 | 0.667 | 0.772 | |
| 5 | -2118.390 | 0.371 | -15.879 | 39.069 | 7.986 | 76.600 | 2.110 | 2.1119 | 2.099 | 2.100 | |
| 7 | -2213.006 | 0.522 | -15.540 | 42.003 | 7.347 | 72.100 | 1.046 | 0.7385 | 0.725 | 0.737 | |
| 8 | -2288.181 | 0.534 | -12.048 | 43.091 | 6.065 | 77.900 | 0.992 | 1.1959 | 1.310 | 1.298 | |
| 9 | -2327.485 | 0.558 | -12.034 | 43.726 | 6.170 | 75.700 | 1.199 | 0.8560 | 0.665 | 0.769 | |
| 11 | -2534.064 | 0.483 | -15.753 | 41.898 | 7.409 | 76.700 | 0.382 | 0.7792 | 0.649 | 0.628 | |
| 12 | -2573.369 | 0.533 | -15.553 | 42.645 | 7.747 | 73.800 | 0.324 | 0.5348 | 0.473 | 0.472 | |
| 13 | -2648.544 | 0.545 | -12.055 | 43.739 | 6.465 | 79.600 | 1.486 | 0.9925 | 1.416 | 1.288 | |
| 14 | -2687.848 | 0.570 | -12.044 | 44.369 | 6.570 | 77.400 | 0.582 | 0.6502 | 0.629 | 0.740 | |
| 15 | -2534.064 | 0.485 | -15.692 | 41.838 | 7.409 | 76.700 | 0.892 | 0.7776 | 0.641 | 0.625 | |
| 16 | -2573.369 | 0.533 | -15.557 | 42.558 | 7.747 | 73.800 | 0.617 | 0.5202 | 0.461 | 0.464 | |
| 18 | -2188.972 | 0.518 | -15.798 | 44.081 | 6.724 | 73.200 | 0.629 | 0.7116 | 0.675 | 0.678 | |
| 19 | -2228.277 | 0.571 | -15.560 | 44.843 | 7.062 | 70.600 | 0.949 | 0.5258 | 0.762 | 0.761 | |
| 20 | -2342.756 | 0.624 | -12.094 | 46.444 | 5.886 | 74.100 | 0.474 | 0.5699 | 0.495 | 0.515 | |
| 21 | -2228.275 | 0.578 | -15.558 | 44.824 | 7.062 | 70.600 | 0.629 | 0.5114 | 0.764 | 0.756 | |
| 22 | -2342.754 | 0.615 | -12.040 | 46.569 | 5.886 | 74.100 | 0.581 | 0.6138 | 0.527 | 0.550 | |
| 23 | -2303.447 | 0.477 | -13.677 | 47.151 | 6.598 | 70.600 | 0.702 | 0.7632 | 0.718 | 0.768 | |
| 25 | -2457.232 | 0.593 | -9.705 | 49.569 | 5.759 | 71.600 | 0.661 | 0.6775 | 0.659 | 0.611 | |
| 26 | -2191.186 | 0.989 | -15.240 | 49.085 | 7.482 | 71.700 | 1.102 | 1.2207 | 1.110 | 1.177 | |
| 27 | -2230.491 | 1.046 | -14.930 | 49.892 | 7.820 | 69.200 | 1.319 | 1.0648 | 1.320 | 1.174 | |
| 28 | -2305.667 | 1.058 | -11.375 | 51.018 | 6.538 | 74.400 | 1.239 | 1.4323 | 1.243 | 1.280 | |
| 30 | -2191.189 | 0.605 | -15.188 | 46.528 | 7.381 | 71.700 | 1.449 | 1.3842 | 1.403 | 1.300 | |
| 31 | -2230.494 | 0.660 | -14.869 | 47.331 | 7.719 | 69.200 | 1.084 | 1.2315 | 1.108 | 1.204 | |
| 32 | -2305.670 | 0.669 | -11.361 | 48.401 | 6.437 | 74.400 | 1.655 | 1.5870 | 1.661 | 1.600 | |
| 34 | -2074.665 | 0.796 | -6.763 | 43.187 | 5.271 | 72.000 | 0.508 | 0.5254 | 0.508 | 0.392 | |
| 35 | -2113.970 | 0.851 | -6.497 | 43.975 | 5.609 | 69.300 | 0.334 | 0.3270 | 0.356 | 0.390 | |
| 37 | -2228.449 | 0.885 | -3.101 | 45.637 | 4.433 | 72.900 | 0.441 | 0.4175 | 0.436 | 0.427 | |
3.1. Multiple linear regression
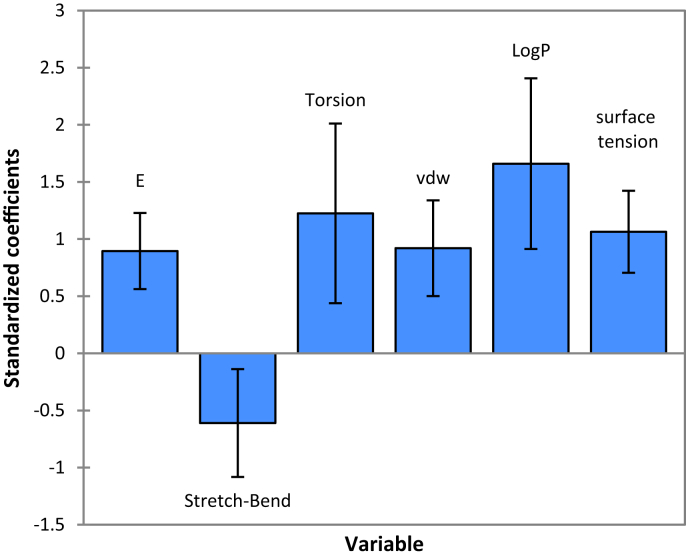
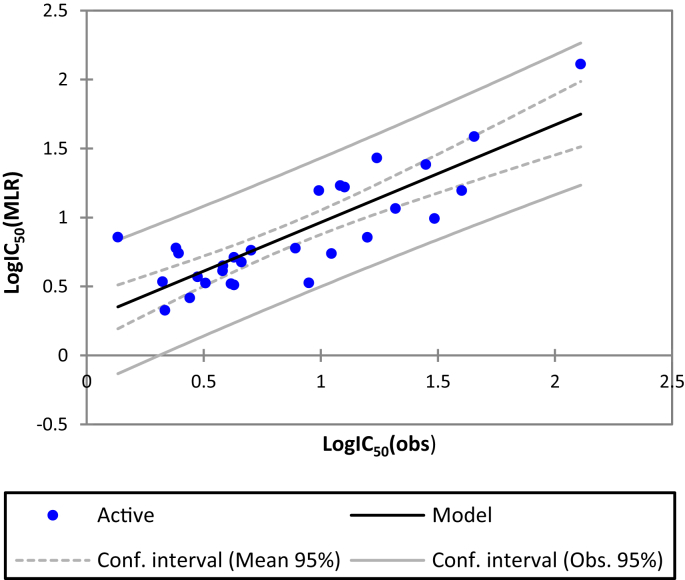
The MLR method is based on three criteria: Coefficient of determination (R2), the root mean square error (RMSE) and the Fisher ratio value (F) [14]. The MLR results which contain the corresponding normalized descriptors coefficients and the correlation between the observed and predicted activities are presented in Figs. 2 and 3 respectively. In addition, the QSAR model of the training set built is represented by the following Eq. (1):
| LogIC50 = -15.8 + 0.0026*E – 1.67* S–B + 0.18*T + 0.15*VDW + 0.91 LogP + 0.18* ST | (1) |
N = 29 R = 0.84 R2 = 0.70 F = 8.84 RMSE = 0.29
Fig. 2.
Modeling characterization by the normalized coefficients.
Fig. 3.
The correlation between the observed and the predicted activities.
The selected descriptors involved in the MLR model of the training set are: Total energy (E), stretch-bend energy (SB), Torsion energy (T), VDW energy (EVDW), lipophilic (LogP) and Surface Tension (ST). According to the coefficient normalization diagram, we have found that the built model presented four most important descriptors (LogP, T, ST and S-b) correlated with Log IC50 with high value of R2 (0.7). Indeed, the most important parameter in the model is LogP (coefficient of 0.91). The positive sign of the Log P Eq. (1), indicates that the larger the Log P value, the lower the activity of the compound is. From Table 3, compounds 4 and 5, which have respectively the lowest and the largest Log P values, also have the greatest and the minor antimalarial activities. The compound 4 possess large alkyl chains; therefore, it is possible that there are hydrophobic interactions between the substituent and the receptor.
The second most important descriptor in Model 1 is the T and ST (coefficient of 0.18). The number of torsion plays a very important role in the activity, this is due to the flexibility of the molecule inside the active site. Moreover, the Surface tension is closely related to the forces of intermolecular attraction. The stronger the intermolecular forces are, the more tightly the molecules are held together in the liquid phase and, therefore the higher the surface tension will be. T and ST appears in the Eq. (1) with apositive sign which shows that the molecules with higher value of T and ST have the lowest antimalarial activity.
The third most important descriptor in model 1 is S–B (coefficient of -1.67). The S–B a geometry parameter, deals with the stretching and bending or one can say the conformational flexibility of the molecule. The descriptor S–B exhibits negative correlation with LogIC50, so the substituents that increase the stretch-bend energy of the compound will also enhance the antimalarial activity.
High correlation coefficient (R = 0.84) of the built QSAR model based on the training set indicates good variance explanation of the model, further supported by low standard deviation (RMSE = 0.23). Furthermore, evaluation of the degree of visual significance of the Fischer Test (F) confidence (p < 0.0001) reflects the good predictive competence of the generated model.
3.2. Artificial neural networks
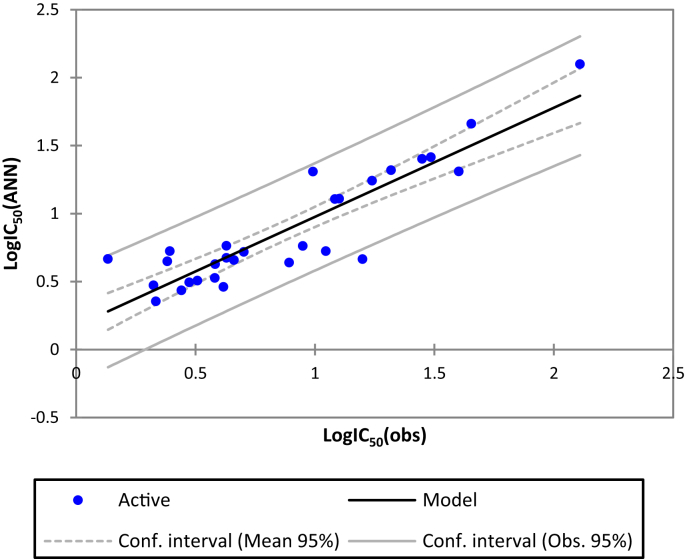
In this study, we have used 3 layers Neural Networks: The input layer that contains six neurons representing the selected descriptors, the output layer which represent the observed activity values (logIC50) and the hidden layer. It should be noted that there are no theoretical or empirical rules to determinate the number of hidden layers. While, few authors [37, 38] recommended to take into consideration ρ= (number of weight)/(number of connection) parameter which must in the range of 1 < ρ < 3 [39,40]. Thus, the final ANN architecture is (6-2-1). The Fig. 4 shows the correlation between the observed and the predicted activities established by the ANN.
Fig. 4.
The correlation between the observed and the predicted activities established by ANN.
The plot of observed versus predicted activity (Fig. 4) shows a good fit with R2 value of 0.81 and the mean squared error RMSE = 0.18 indicate that model possesses a significant statistical quality and the selected descriptors by MLR are pertinent.
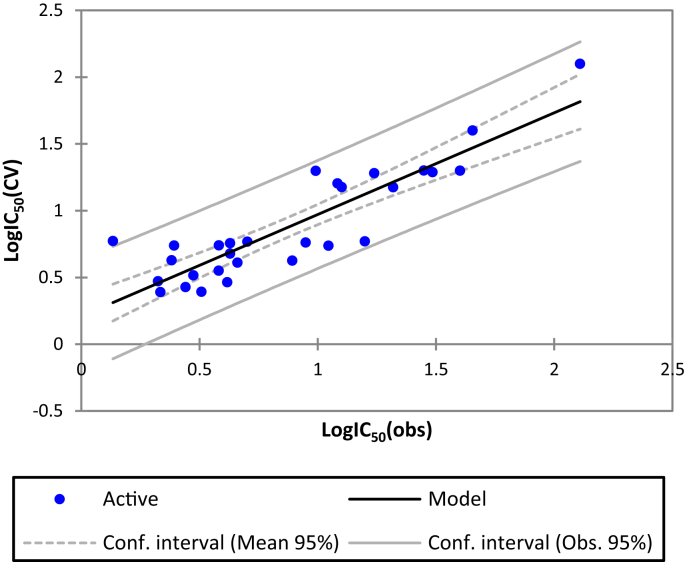
3.3. Cross Validation
The results obtained by Cross Validation (CV)with “leave-one-out”are represented in Fig. 5. Therefore, the obtained parameters R2 = 0.78 and RMSE = 0.19 shows that the built QSAR model is not sensitive to the CV. Which obviously, indicate that the proposed QSAR model is stable and robust. However, Cross Validation is not a good parameter to estimate the ability of QSAR models according to Golbraikh and Tropsha study [31].
Fig. 5.
Correlation of observed and predicted activities calculated using Cross-Validation (LOO).
3.4. Y-randomization
Herein, we have randomly mixed descriptors and observed activity of the newly training set which contains 29 compounds obtained by excluding 8 compounds and adding 8 compounds of test set. Then, we build new model through MLR methods us described previously. The obtained results are given in Table 4.
Table 4.
Calculated results using Y-randomization.
| N | 4 | 23 | 5 | 26 | 30 | 37 | 2 | 32 | 24 | 21 | 28 | 9 | 7 | 20 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| logIC50 | 0.134 | 0.702 | 2.110 | 1.102 | 1.449 | 0.441 | 0.393 | 1.655 | 0.412 | 0.629 | 1.239 | 1.199 | 1.046 | 0.474 | 0.382 |
| pred logIC50 | 0.869 | 0.732 | 2.102 | 1.254 | 1.368 | 0.401 | 0.774 | 1.534 | 0.588 | 0.543 | 1.428 | 0.867 | 0.772 | 0.585 | 0.802 |
| N | 36 | 1 | 27 | 25 | 16 | 19 | 13 | 31 | 8 | 12 | 34 | 17 | 33 | 15 | - |
| logIC50 | 0.769 | 1.358 | 1.319 | 0.661 | 0.617 | 0.949 | 1.486 | 1.084 | 0.992 | 0.324 | 0.508 | 0.666 | 1.308 | 0.892 | - |
| pred logIC50 | 0.716 | 1.032 | 1.080 | 0.610 | 0.528 | 0.556 | 0.980 | 1.196 | 1.211 | 0.541 | 0.545 | 0.626 | 1.261 | 0.800 | - |
The newly QSAR model using the Y-randomization method is represented by the following Eq. (2):
| LogIC50 = -15.08 + 0.0026*E - 1.44*S–B +0.16*T + 00.14*VDW + 0.85*LogP + 0.17*ST | (2) |
N = 29 R = 0.83 R2 = 0.69 F = 8.14 RMSE = 0.29
The obtained correlation coefficient value R2 = 0.69 of the newly training set is compared to the obtained using the first training set. The results of Y-randomization confirm the absence of dependence between descriptors included in the QSAR model.
3.5. External validation
The study conducted by Golbraikh and Tropsha on validation methods lead to the insufficient of the internal validation methods to confirm the reliability of the built QSAR models. For this reason, external validations are hardly needed to build a reliable QSAR model [31]. The later must respect some important criteria recommended by Golbraikh and Tropsha. In Table 5, we present the test set containing 8 compounds that has been reserved to external validation, maintaining their original numbers and taken from Table 3 with their observed and predicted activity by MLR and ANN models. The results of Golbraikh and Tropsha criteria's validation are presented in Table 6 [31].
Table 5.
The results of external validation by the MLR and ANN methods.
| N | LogIC50 | Pred (LogIC50) MLR | Residual | Pred (LogIC50) | Residual |
|---|---|---|---|---|---|
| 1 | 1.358 | 0.99 | 0.388 | 0.776 | 0.443 |
| 6 | 0.953 | 0.984 | -0.013 | 0.769 | 0.042 |
| 10 | 1.621 | 2.119 | -0.158 | 2.099 | -0.109 |
| 17 | 0.666 | 0.65 | -0.061 | 0.611 | -0.146 |
| 24 | 0.412 | 0.646 | -0.312 | 0.525 | -0.347 |
| 29 | 1.167 | 1.155 | 0.079 | 1.014 | 0.106 |
| 33 | 1.308 | 1.326 | 0.097 | 1.393 | 0.013 |
| 36 | 0.769 | 0.736 | -0.02 | 0.544 | -0.002 |
Table 6.
Golbraikh and Tropsha criteria.
| Parameter | Formula | Threshold | Modelscore |
|---|---|---|---|
| >0.6 | 0.68 | ||
| r2 | Coefficient of determination for the plot of predicted versus observed for test set by MLR | r2 > 0.6 | 0.74 |
| r2 at zero intercept | 0.67 | ||
| r2 for the plot of observed versus predicted activity for the test set at zero intercept | 0.68 | ||
| 0.01 | |||
| k | Slope of the plot of predicted versus observed activity for test set at zero intercept | 0.85 < k < 1.15 | 0.92 |
| 0.07 | |||
| k’ | Slope of the plot of observed versus predicted activity at zero intercept | 0.85 < k’<1.15 | 0.93 |
| 0.08 |
Overall, we can conclude that Golbraikh and Tropsha criteria's and external validation are successfully validated, which indicate that the built QSAR model is in perfect agreement with all validation methods in one hand. In the other hand, the experimental antimalarial activity could be accurately predicted using the established QSAR model.
3.6. Molecular docking study
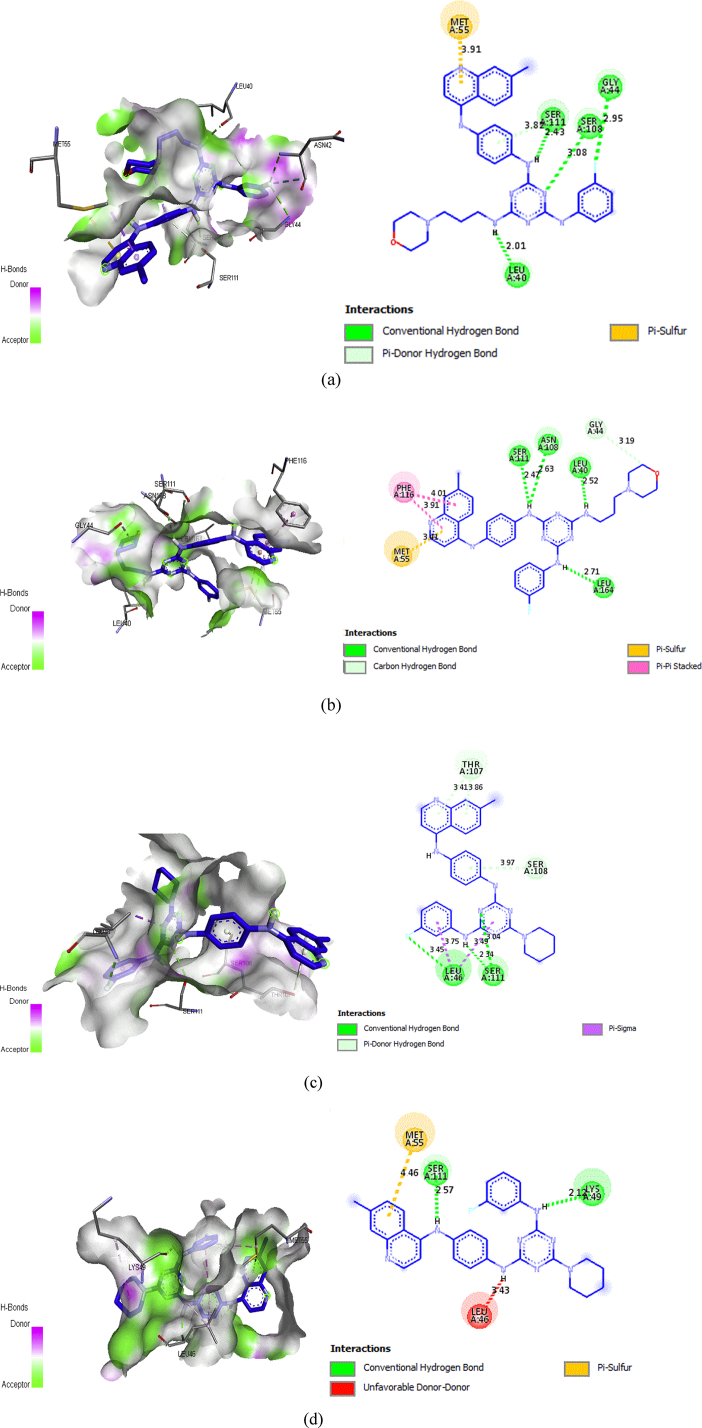
The molecular Docking study was performed for two reasons. The first is to understand the good antimalarial activity potency manifested with some compounds. The second is to find out the key interaction types established with the protein (pf-DHFR) in its two type (wild and mutant) [14]. The reported study of Yuvanyama et al [41] has found the binding modes, and has localized the active sites in wild and mutant of protein (pf-DHFR). The study performed with a potent inhibitor 1,3,5-triazine derivative which is a preclinical molecule called WR99210. It is found that the important sites in the case of the wild type are located in Ile14, Ala16, Met55, Asp54, Ser108, Ile164 and Tyr170. It was also found that important sites are located in Ala16, Cys50, Asn51, Cys59, Asn108, Leu164 and Tyr170 in the case of the wild type. The interactions mode obtained by molecular docking for compounds 4 and 5 are presented in Fig. 6.
Fig. 6.
2D and 3D docking poses showing interactions of compounds 4 and5 in the binding sites of wild type and quadruple mutant of pf-DHFR-TS. (a) Compound 4: wild type of pf-DHFR (binding energy −10.6 kcal/mol). (b) Compound 4: quadruple mutant of pf- DHFR-TS (binding energy −10.9 kcal/mol). (c) Compound 5: wild type of pf-DHFR (binding energy −11.4 kcal/mol). (d) Compound 5: quadruple mutant of pf-DHFR-TS (binding energy −10.3 kcal/mol).
In the case of the wild type, compound 4 performs four interactions through hydrogen bonding with SER111, LEU40, SER108 and GLY44 amino acids. The interactions involve three nitrogen atoms linked to the triazine system and one fluorine atom linked to benzene group, with the distance of 2.43 Ǻ, 2.01 Ǻ, 3.08 Ǻ and 2.95 Ǻ, respectively. However, compound 5 performs only three hydrogen bonds with less important binding sites, Ser111 and Gly46, as they are not cited as active sites for antimalarial activity. In the case of quadruple mutant, compound 4performs four hydrogen bonds between four nitrogenatoms linked to the triazine group and SER111, ASN108, LEU164 and LEU40 amino acids with the distance of 2.47 Ǻ, 2.63 Ǻ, 2.71 Ǻ and 2.52 Ǻ, respectively. For compound 5 showed only two hydrogen bonding interaction with less important binding sites SER 111 and LYS49.
In summary, the interactions formed by the compounds 4 with the binding sites of pf-DHFR are in good agreement with the previous study for antimalarial activity [41]. Moreover, no significant interactions with critical amino acid in pf-DHFR protein on both wild and mutant types, showed for compound 5. When, we have noticed for compound 4 bearing N-methy-3-morpholinopropan-1-amine substitute, the number of rotations increased 11 rotations versus 7 rotations in the case of compound 5. This could make molecule more flexible inside the active site of the protein. Furthermore, the introduction of new nitrogen atoms in the compound 4 influence on the number of hydrogen bond compared to compound 5. These outcomes, can explain the difference of measured activity between compound 4 and compound 5.
4. Conclusion
In the purpose of producing new effective antimalarial drugs, a QSAR model was developed using the in vitro antimalarial data reported. The built model was statistically significant and the significance was validated. Molecular docking study highlight the exclusive binding signature of the ligands with the active site residue i.e. ILE164, SER108 and LEU40of the target and it explains the specificity and subtle differences in their predicted IC50 values. The study has provided insights to improve biological activity with the change of 1-methylpiperidine by 3-morpholinopropan-1-aminein 4-aminoquinoline-triazinederivatives.
Declarations
Author contribution statement
Hanine Hadni: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Menana Elhallaoui: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Vaishnani J.B. Cutaneous findings in five cases of malaria. Indian J. Dermatol. Venereol. Leprol. 2011;77:110. doi: 10.4103/0378-6323.74985. [DOI] [PubMed] [Google Scholar]
- 2.Murray M.C., Perkins M.E. Chapter 15. Chemotherapy of malaria. Annu. Rep. Med. Chem. 1996;31:141–150. [Google Scholar]
- 3.White N.J., Pukrittayakamee S., Hien T.T., Faiz M.A., Mokuolu O.A., Dondorp A.M. Malaria. Lancet. 2014;383:723–735. doi: 10.1016/S0140-6736(13)60024-0. [DOI] [PubMed] [Google Scholar]
- 4.Newton P., White N. MALARIA: new developments in treatment and prevention. Annu. Rev. Med. 1999;50:179–192. doi: 10.1146/annurev.med.50.1.179. [DOI] [PubMed] [Google Scholar]
- 5.WHO | Global Malaria Mapper. WHO; 2016. http://www.who.int/malaria/publications/world_malaria_report/global_malaria_mapper/en/ [Google Scholar]
- 6.Brock A.R., Ross J.V., Parikh S., Esterman A. The role of antimalarial quality in the emergence and transmission of resistance. Med. Hypotheses. 2018;111:49–54. doi: 10.1016/j.mehy.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Prosser C., Meyer W., Ellis J., Lee R. Evolutionary ARMS race: antimalarial resistance molecular surveillance. Trends Parasitol. 2018;34:322–334. doi: 10.1016/j.pt.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Muregi F.W., Ishih A. Next-generation antimalarial drugs: hybrid molecules as a new strategy in drug design. Drug Dev. Res. 2009;71 doi: 10.1002/ddr.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hastings M.D., Sibley C.H. Pyrimethamine and WR99210 exert opposing selection on dihydrofolate reductase from Plasmodium vivax. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13137–13141. doi: 10.1073/pnas.182295999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boechat N., Ferreira M. de L.G., Pinheiro L.C.S., Jesus A.M.L., Leite M.M.M., Júnior C.C.S., Aguiar A.C.C., de Andrade I.M., Krettli A.U. New compounds hybrids 1 H -1,2,3-Triazole-Quinoline against Plasmodium falciparum. Chem. Biol. Drug Des. 2014;84:325–332. doi: 10.1111/cbdd.12321. [DOI] [PubMed] [Google Scholar]
- 11.Mishra A., Batchu H., Srivastava K., Singh P., Shukla P.K., Batra S. Synthesis and evaluation of new diaryl ether and quinoline hybrids as potential antiplasmodial and antimicrobial agents. Bioorg. Med. Chem. Lett. 2014;24:1719–1723. doi: 10.1016/j.bmcl.2014.02.044. [DOI] [PubMed] [Google Scholar]
- 12.Roy K., Kar S., Das R.N. SAR and QSAR in drug discovery and chemical design—some examples. Underst. Basics QSAR Appl. Pharm. Sci. Risk Assess. 2015 [Google Scholar]
- 13.Sarvagalla S., Syed S.B., Coumar M.S. An overview of computational methods, tools, servers, and databases for drug repurposing. Silico Drug Des. 2019 [Google Scholar]
- 14.Hadni H., Mazigh M., Charif E., Bouayad A., Elhallaoui M. Molecular modeling of antimalarial agents by 3D-QSAR study and molecular docking of two hybrids 4-Aminoquinoline-1,3,5-triazine and 4-Aminoquinoline-oxalamide derivatives with the receptor protein in its both wild and mutant types. Biochem. Res. Int. 2018:1–15. doi: 10.1155/2018/8639173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hadni H., Mazigh M., Elhallaoui M. QSAR and Molecular docking studies of 4-anilinoquinoline- triazine hybrids as pf-DHFR inhibitors. 2019;8:84–93. doi: 10.1016/j.heliyon.2019.e02357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma M., Chauhan K., Chauhan S.S., Kumar A., Singh S.V., Saxena J.K., Agarwal P., Srivastava K., Raja Kumar S., Puri S.K., Shah P., Siddiqi M.I., Chauhan P.M.S. Synthesis of hybrid 4-anilinoquinoline triazines as potent antimalarial agents, their in silico modeling and bioevaluation as Plasmodium falciparum transketolase and ??-hematin inhibitors. MedChemComm. 2012;3:71–79. [Google Scholar]
- 17.Lill M.A. Multi-dimensional QSAR in drug discovery. Drug Discov. Today. 2007;12:1013–1017. doi: 10.1016/j.drudis.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Du Q.-S., Gao J., Wei Y.-T., Du L.-Q., Wang S.-Q., Huang R.-B. Structure-based and multiple potential three-dimensional quantitative structure–activity relationship (SB-MP-3D-QSAR) for inhibitor design. J. Chem. Inf. Model. 2012;52:996–1004. doi: 10.1021/ci300066y. [DOI] [PubMed] [Google Scholar]
- 19.Hoekman D. Exploring QSAR fundamentals and applications in chemistry and biology, volume 1. Hydrophobic, electronic and steric constants, volume 2. J. Am. Chem. Soc. 1995;117:9782. [Google Scholar]
- 20.Sirawaraporn W., Sathitkul T., Sirawaraporn R., Yuthavong Y., V Santi D. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1124–1129. doi: 10.1073/pnas.94.4.1124. http://www.ncbi.nlm.nih.gov/pubmed/9037017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Österberg T., Norinder U. Prediction of drug transport processes using simple parameters and PLS statistics the use of ACD/logP and ACD/ChemSketch descriptors. Eur. J. Pharm. Sci. 2001;12:327–337. doi: 10.1016/s0928-0987(00)00189-5. [DOI] [PubMed] [Google Scholar]
- 22.Milne G.W.A. Software Review of ChemBioDraw 12.0 ChemBioDraw 12.0 CambridgeSoft 100 CambridgePark Drive, Cambridge, MA 02140, United States. J. Chem. Inf. Model. 2010;50:2053. http://www.cambridgesoft.com/software/ChemBioDraw/ [Google Scholar]
- 23.Parr R.G., Yang W. 1989. Density-Functional Theory of Atoms and Molecules. [Google Scholar]
- 24.Becke A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993;98:5648–5652. [Google Scholar]
- 25.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Montgomery J.A., Vreven T., Kudin K.N., Burant J.C., Millam J.M., Iyengar S.S., Tomasi J., Barone V., Mennucci B., Cossi M., Scalmani G., Rega N., Petersson G.A., Nakatsuji H., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Klene M., Li X., Knox J.E., Hratchian H.P., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Ayala P.Y., Morokuma K., Voth G.A., Salvador P., Dannenberg J.J., Zakrzewski V.G., Dapprich S., Daniels A.D., Strain M.C., Farkas O., Malick D.K., Rabuck A.D., Raghavachari K., Foresman J.B., V Ortiz J., Cui Q., Baboul A.G., Clifford S., Cioslowski J., Stefanov B.B., Liu G., Liashenko A., Piskorz P., Komaromi I., Martin R.L., Fox D.J., Keith T., Al-Laham M.A., Peng C.Y., Nanayakkara A., Challacombe M., Gill P.M.W., Johnson B., Chen W., Wong M.W., Gonzalez C., Pople J.A. Gaussian 03, revision C. 2004;02 [Google Scholar]
- 26.Chtita S., Ghamali M., Ousaa A., Aouidate A., Belhassan A., Taourati A.I., Masand V.H., Bouachrine M., Lakhlifi T. QSAR study of anti-Human African Trypanosomiasis activity for 2-phenylimidazopyridines derivatives using DFT and Lipinski’s descriptors. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e01304. e01304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta M.K., Gupta S., Rawal R.K. Artif. Neural Netw. Drug Des. Deliv. Dispos. Elsevier; 2016. Impact of artificial neural networks in QSAR and computational modeling; pp. 153–179. [Google Scholar]
- 28.Flood I., Kartam N. Neural networks in civil engineering. II: systems and application. J. Comput. Civ. Eng. 1994;8:149–162. [Google Scholar]
- 29.Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. J. Am. Stat. Assoc. 1983;78:316–331. [Google Scholar]
- 30.Stone M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. 1974 [Google Scholar]
- 31.Golbraikh A., Tropsha A. Beware of q2! J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- 32.Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol. Inform. 2010;29:476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 33.Kouranov A., Xie L., de la Cruz J., Chen L., Westbrook J., Bourne P.E., Berman H.M. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302. doi: 10.1093/nar/gkj120. –D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.BIOvIA D.S. Vol. 4. Dassault Syst. Release; San Diego: 2015. Discovery Studio Modeling Environment. [Google Scholar]
- 35.Morris G., Huey R. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 37.So S.S., Richards W.G. Application of neural networks: quantitative structure-activity relationships of the derivatives of 2,4-diamino-5-(substituted-benzyl)pyrimidines as DHFR inhibitors. J. Med. Chem. 1992;35:3201–3207. doi: 10.1021/jm00095a016. [DOI] [PubMed] [Google Scholar]
- 38.Andrea T.A., Kalayeh H. Applications of neural networks in quantitative structure-activity relationships of dihydrofolate reductase inhibitors. J. Med. Chem. 1991;34:2824–2836. doi: 10.1021/jm00113a022. [DOI] [PubMed] [Google Scholar]
- 39.Naceiri Mrabti M.E.N., Dguigui K., Hadni H. QSAR study and molecular docking of benzimidazole derivatives as potent activators of AMP-activated protein kinase. J. Taibah Univ. Sci. 2017;11:18–39. [Google Scholar]
- 40.Kůrková V. Kolmogorov’s theorem and multilayer neural networks. Neural Netw. 1992 [Google Scholar]
- 41.Yuvaniyama J., Chitnumsub P., Kamchonwongpaisan S., Vanichtanankul J., Sirawaraporn W., Taylor P., Walkinshaw M.D., Yuthavong Y. Insights into antifolate resistance from malarial DHFR-TS structures. Nat. Struct. Biol. 2003;10:357–365. doi: 10.1038/nsb921. [DOI] [PubMed] [Google Scholar]