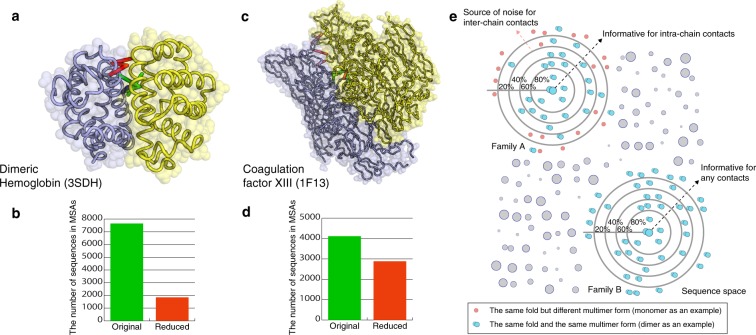
Figure 4.
A practical example of our method and enhanced CSs in contact pairs of homodimers in reduced MSAs. (a) Enhanced CSs in contact pairs of homodimeric hemoglobin. Red bars show contact pairs with CS higher than 0.6 when using the reduced alignment. Blue bars show those when using the original alignment. Green bars show contact pairs having CS higher than 0.6 in both alignments. (b) Number of sequences included in MSAs of 3SDH with the default (green) and the conservative (red) thresholds. (c) Enhanced CSs in contact pairs of human coagulation factor XIII. Color scheme is the same as 3SDH. (d) Number of sequences included in MSAs of 1F13. Color scheme is the same as 3SDH. (e) Schematic diagram of differences in contact prediction of interchain and intrachain interactions. Required sequences for the estimation of CS depend on the degree of conservation in target contacts. In general, CS estimation of interchain contacts requires more similar sequences because of their lower degree of conservation of oligomeric states compared to folds. Although intrachain contacts are more conserved (i.e., even quite diverged sequences are still informative for intrachain contact estimation), interchain contacts are less conserved because oligomeric states can vary more than folds. This is at least true for our manually confirmed samples. In such cases, greatly diverged sequences, which accommodate different (oligomeric) states from that of the target, can be a source of noise in the estimation. The sequence threshold for these samples is apparently appropriate at 20–40%. Family A in the figure illustrates a protein family where diverged sequences can be noise. In contrast, even diverged sequences are still informative for interchain contacts if oligomeric state is highly conserved. Family B is an example for this case.