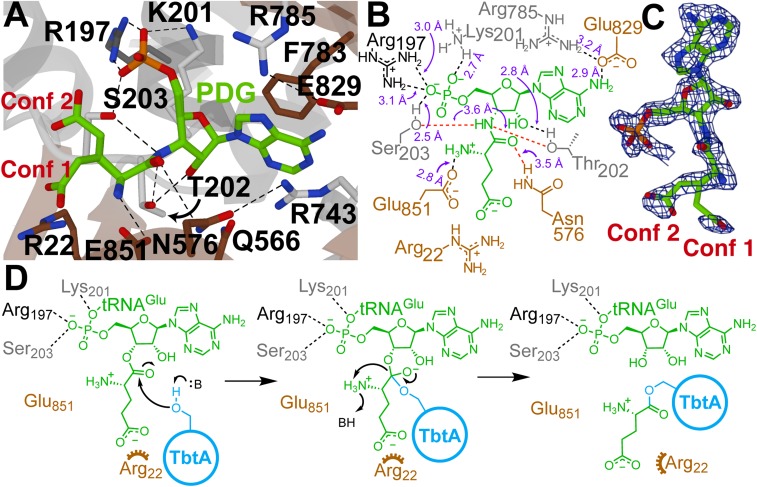
Fig. 3.
Structural analysis of the active site of TbtB. (A) TbtB bound to PDG (2.15 Å; PDB 6EC8) with active-site residues targeted for mutagenesis represented as sticks (residues residing in an α-helix, β-strand, or random coil are shown in dark gray, brown, or light gray, respectively). (B) Two-dimensional representation of A with labeled interatomic distances (≤3.2 Å in black; ≥3.5 Å in red). The distances reflect crystallographic nonhydrogen interatomic distances, but the dashed lines are drawn to satisfy proposed hydrogen-bonding interactions. (C) Feature-enhanced map contoured at 2σ for the PDG ligand (30). PDG was modeled as 2 conformations [Conf 1 (10%) and Conf 2 (90%)] due to partial disorder of the Glu side-chain atoms. (D) Proposed mechanism of TbtB-catalyzed glutamylation of TbtA. An alternative ping-pong mechanism, described in the text, was ruled out based on site-directed mutagenesis studies.