Significance
Cytomegalovirus is a herpesvirus that persists latently for the host’s lifetime, with periodic reactivation that can cause life threatening disease in individuals with inadequate immunity. Understanding the mechanisms by which the virus reactivates is critical to controlling reactivation and disease. For decades, reactivation of viral gene expression and replication has been presumed to be driven from de-repression of the major immediate early promoter (MIEP). We show that viral reactivation depends on alternative promoter elements within the major immediate early gene locus while the MIEP largely remains silent during reactivation. This finding represents a major paradigm shift, demonstrating the use of alternative promoters to drive virus reactivation in a context-dependent manner.
Keywords: human cytomegalovirus, latency, reactivation
Abstract
Reactivation from latency requires reinitiation of viral gene expression and culminates in the production of infectious progeny. The major immediate early promoter (MIEP) of human cytomegalovirus (HCMV) drives the expression of crucial lytic cycle transactivators but is silenced during latency in hematopoietic progenitor cells (HPCs). Because the MIEP has poor activity in HPCs, it is unclear how viral transactivators are expressed during reactivation. It has been presumed that viral gene expression is reinitiated via de-repression of the MIEP. We demonstrate that immediate early transcripts arising from reactivation originate predominantly from alternative promoters within the canonical major immediate early locus. Disruption of these intronic promoters results in striking defects in re-expression of viral genes and viral genome replication in the THP-1 latency model. Furthermore, we show that these promoters are necessary for efficient reactivation in primary CD34+ HPCs. Our findings shift the paradigm for HCMV reactivation by demonstrating that promoter switching governs reactivation from viral latency in a context-specific manner.
Reactivation of latent human cytomegalovirus (HCMV) infection poses a life-threatening risk to immunocompromised individuals, such as stem cell or organ transplant recipients (1). While the HCMV replicative cycle has been studied extensively, our understanding of the mechanisms controlling the entry into and exit from latency is far from complete.
During productive infection, the HCMV genome is transcribed in a temporal cascade composed of three kinetic classes of gene expression designated as immediate early (IE), early, and late (2). The IE proteins, in particular IE1-72 kilodalton (kDa) and IE2-86 kDa proteins, referred to as IE1 and IE2, play critical roles in initiating the HCMV lytic cycle by transactivating the expression of cellular and viral genes and suppressing the innate immune response (3). During latency, IE gene expression is repressed, resulting in diminished viral gene expression and the absence of productive replication (4). Signals that stimulate reactivation induce IE gene expression to allow reentry into the viral replicative cycle (5), and this de-repression of IE genes is considered a pivotal event controlling the switch between latent and reactivated states.
The major immediate early promoter (MIEP) is a powerful promoter in cells permissive for lytic HCMV replication and drives high level expression of mRNAs encoding IE1 (UL123) and IE2 (UL122) (6–9). In cell types that support HCMV latency, however, such as CD34+ human progenitor cells (HPCs) and CD14+ monocytes, MIEP activity is diminished (10–12). Because re-expression of UL122 and UL123 is required for reactivation (13–15), it has been presumed that de-repression of the MIEP is critical to reinitiate the viral lytic cycle. However, the origin of UL123 and UL122 transcripts during HCMV reactivation has not been formally defined.
Results
Re-Expression of UL123 (IE1) and UL122 (IE2) following Reactivation Stimulus Does Not Originate from the MIEP.
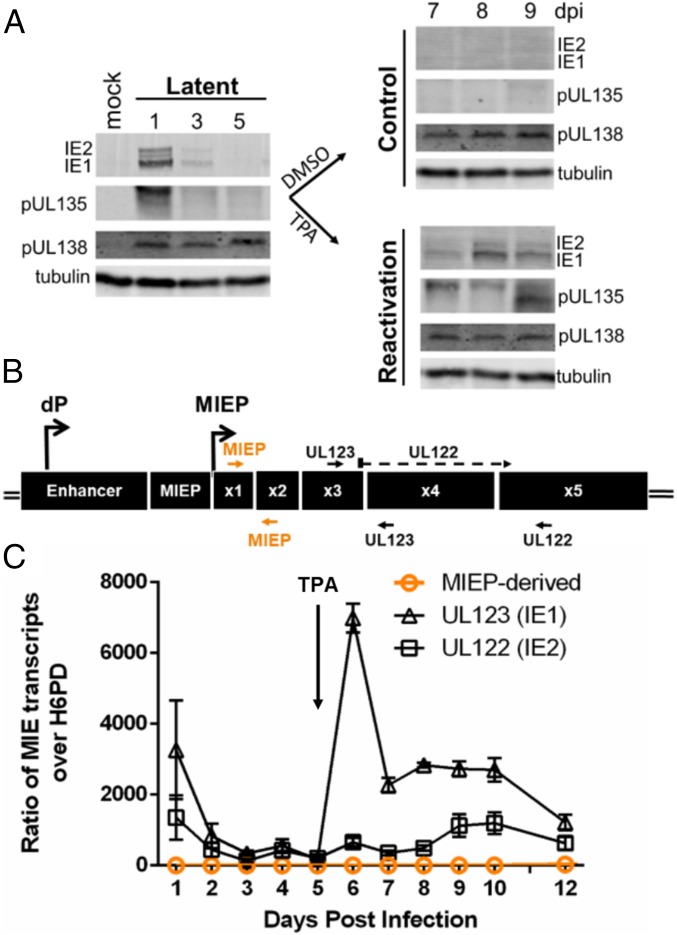
We measured the accumulation of IE1 and IE2 proteins and the expression of UL123 and UL122 transcripts during experimental latency and reactivation in the monocytic THP-1 cell line. THP-1 cells are an established model for studying HCMV latency and reactivation (16–19). While the THP-1 cell model does not recapitulate all aspects of latency, such as the robust production of progeny virus, the THP-1 cell line offers the strength of a synchronous reactivation. Cells were infected with HCMV (TB40/E strain) and allowed to establish latency for 5 d post infection (dpi). To induce reactivation, latently infected cells were treated with 12-O-tetradecanoylphorbol-13-acetate (TPA), which promotes monocyte-to-macrophage differentiation and triggers viral reactivation (16, 17). IE1 and IE2 proteins were detected immediately after infection but decreased to levels below the limit of detection between 2 and 5 dpi (Fig. 1A and the more detailed time course in SI Appendix, Fig. S1), consistent with the establishment of latency. IE1 and IE2 proteins accumulated rapidly following TPA treatment, consistent with viral reactivation. Parallel cultures treated with the solvent control (dimethyl sulfoxide, DMSO) maintained latency, and levels of IE1 and IE2 proteins remained below the limit of detection. The viral protein pUL138 (a suppressor of viral replication that promotes latency) (20, 21) is detected at all time points. pUL135 (which promotes reactivation and viral replication) (21) is detected at 1 dpi and then re-expressed following the reactivation stimulus, similar to the IE proteins.
Fig. 1.
IE1 and IE2 are expressed during HCMV reactivation but do not arise from the MIEP. (A) THP-1 cells were infected with TB40/E WT HCMV (MOI = 2) and cultured for 5 d to allow the establishment of latency. At day 5, cells were treated with TPA or a DMSO control. Whole-cell lysates were collected at the indicated time points, and IE1, IE2, pUL135, and pUL138 viral proteins were detected by immunoblotting. Tubulin was used as a loading control. A single experiment (representative of 3 independent experiments) is shown. (B) Schematic of the major immediate early (MIE) locus. The distal promoter (dP) and major immediate early promoter (MIEP) and primers to detect UL123 (IE1) (exons 3 and 4), UL122 (IE2) (spanning exons 3 to 5), or MIEP/dP-derived transcripts (exons 1 and 2, orange) are indicated. (C) UL123 (IE1), UL122 (IE2), and MIEP/dP-derived transcripts were detected by RT-qPCR over a time course following infection and reactivation in THP-1 cells. Transcripts are quantified as a ratio over H6PD. Data from 3 independent biological replicates (each performed in triplicate) are shown; error bars indicate SE.
The accumulation of UL123 and UL122 transcripts following infection and reactivation in THP-1 cells was measured by quantitative reverse transcriptase PCR (RT-qPCR) using primers that detect differentially spliced UL123 and UL122 transcripts (Fig. 1B, black arrows). The expression of UL123 and UL122 transcripts corresponded to protein levels at 1 dpi and following reactivation (Fig. 1C). Surprisingly, a primer pair that detects transcripts arising from the MIEP (transcript UTR136) (22) or the 5′ distal promoter (dP) (23) (transcript UTR406) (22) (Fig. 1B, orange arrows) detected few MIEP/dP-derived transcripts at any time after infection or following reactivation (Fig. 1C). By contrast, the same primer pair readily detected abundant MIEP-derived transcripts during productive infection in fibroblasts (SI Appendix, Fig. S2). The absence of MIEP-derived transcripts following reactivation stimulus in THP-1 cells suggests that the re-expression of IE1 and IE2 is driven by alternative promoters.
Transcripts Derived from MIE Intronic Promoters Predominate following Reactivation Stimulus.
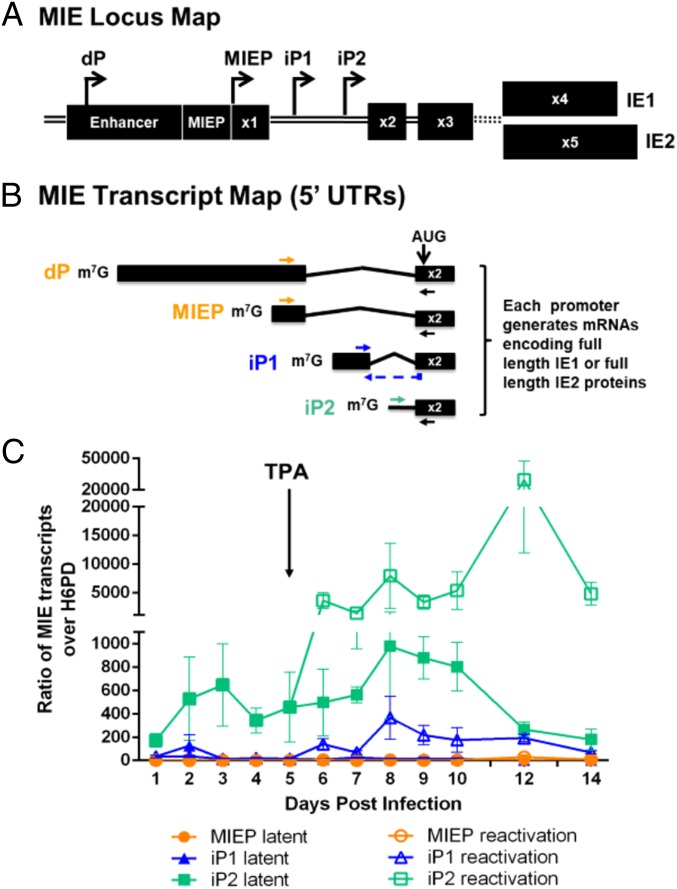
We recently identified two promoters within an intron 3′ of the MIEP [intronic promoter 1 (iP1) and intronic promoter 2 (iP2)] that drive the expression of alternate transcripts (UTR378 and UTR70, respectively) encoding IE1 and IE2 proteins (Fig. 2 A and B) (22). The iP-derived transcripts lack noncoding exon 1 found in MIEP-derived mRNAs but encode the complete set of distal exons necessary to synthesize full-length IE1 or IE2 proteins. As the intronic promoters are dispensable for viral replication in fibroblasts, and transcripts derived from these promoters are expressed with late kinetics during the replicative infection (22) (SI Appendix, Fig. S2), we hypothesized that the iP1 and iP2 might contribute to re-expression of UL122 and UL123 subsequent to a reactivation stimulus.
Fig. 2.
UL123 (IE1) and UL122 (IE2) transcripts derived from intronic promoters predominate following reactivation stimulus. (A) Schematic of MIE locus. The distal promoter (dP), the major immediate early promoter (MIEP), intronic promoter 1 (iP1), and intronic promoter 2 (iP2) give rise to transcripts encoding full-length IE1 and IE2. Transcription start sites are depicted with arrows. (B) Schematic of the 5′ ends up to and including exon 2 of IE-encoding transcripts. The translation start site (AUG) is marked in exon 2. Mature mRNAs encoding IE1 and IE2 will also include exons 3 and 4 or 3 and 5, respectively. Primer pairs designed to detect discrete transcripts by RT-qPCR (dP/MIEP: orange; iP1: blue; iP2: teal) are shown. A common reverse primer was used to amplify dP/MIEP- and iP2-derived transcripts. (C) THP-1 cells were infected with TB40/E WT (MOI = 2) for 5 d to promote latency. At day 5, cells were treated with TPA or DMSO control. The transcripts indicated were quantified relative to H6PD by RT-qPCR. Data from 3 independent biological replicates (each performed in triplicate) are shown; error bars represent SE. Statistical significance was determined by unpaired t test comparing accumulation of each transcript during latency versus reactivation. Welch’s correction was used to account for unequal variance (*P value ≤ 0.05).
We monitored the accumulation of iP1- and iP2-derived transcripts in THP-1 cells using primer pairs that discriminate between the distinct MIE locus transcripts (Fig. 2B; sequences in SI Appendix, Table S1). MIE locus transcripts were predominately derived from iP2 between 1 and 5 dpi, and iP2 transcripts were robustly induced following TPA treatment (Fig. 2C and SI Appendix, Fig. S3). The iP1-derived transcripts also increased after reactivation stimulus, albeit to a lower level. Although a small number of MIEP-derived transcripts were detected from 10 to 14 d following re-expression of MIE genes, MIEP-derived transcripts were far less abundant than those derived from either of the intronic promoters (SI Appendix, Fig. S3). Interestingly, iP-derived transcripts were expressed with later kinetics in fibroblasts despite robust accumulation of MIEP-derived transcripts (SI Appendix, Fig. S2), indicating context-dependent regulation of this locus. As transcripts originating from iP1 and iP2 account for the overwhelming majority of transcripts encoding IE1 and IE2 in THP-1 cells, these findings suggest that the intronic promoters play important roles in the re-expression of IE genes (Fig. 1A) and hence in reactivation from latency.
Intronic Promoter Mutant Viruses Do Not Express IE1 and IE2 following Reactivation Stimulus.
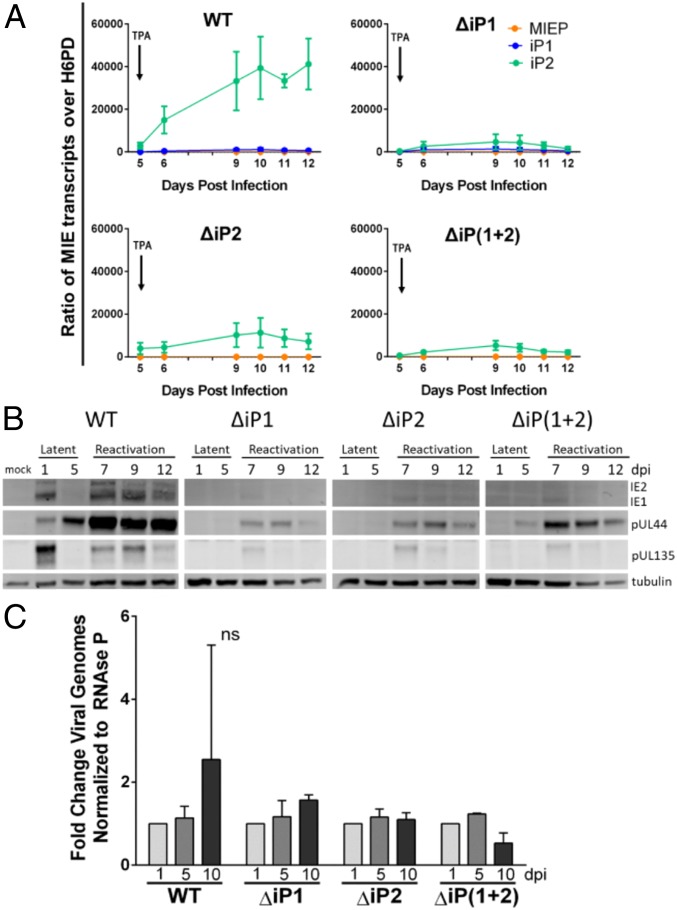
To determine the significance of iP1 and iP2 for viral reactivation, we constructed recombinant viruses containing deletions surrounding the transcription start sites of iP1 (∆iP1), iP2 (∆iP2), or both iP1 and iP2 [∆iP(1 + 2)] (SI Appendix, Fig. S4A). Each mutant virus replicated with similar kinetics and to similar peak titers as the parental wild-type (WT) virus (SI Appendix, Fig. S4B) and efficiently accumulated IE1 and IE2 (SI Appendix, Fig. S4C) during productive infection in fibroblasts. Together, these data indicate that the intronic MIE locus promoters are dispensable in fibroblasts for expression of lytic cycle transactivators and for virus replication.
In THP-1 cells, both iP1- and iP2-derived transcripts accumulated poorly following reactivation stimulus during ∆iP1 infection (Fig. 3A). The iP2-derived transcripts were diminished following reactivation stimulus in ∆iP2 infection; however, iP1 transcript levels could not be measured as the ΔiP2 mutation abolishes the primer binding site for detecting iP1-derived transcripts. Deletion of both iP1 and iP2 similarly diminished levels of iP2-derived transcripts following reactivation stimulus. MIEP-derived transcripts were detected at very low levels, if at all, in all infections. Consistent with the defect in iP-derived mRNA levels, deletion of iP1 and/or iP2 resulted in a striking defect in the accumulation of IE proteins following the reactivation stimulus (Fig. 3B). The ΔiP mutant virus infection also accumulated reduced levels of the early viral proteins pUL44 and pUL135, suggesting a global defect in re-expression of viral gene expression. The failure of the recombinant viruses to reactivate could not be attributed to a defect in maintenance of latent infection, as viral genomes were maintained from 1 to 5 dpi with each virus (Fig. 3C). From these results, we conclude that ΔiP-mutant viruses establish a latent infection but are unable to re-express viral lytic cycle transactivators or representative viral early genes in response to a canonical reactivation stimulus.
Fig. 3.
The iP mutant viruses fail to express IE1 and IE2 following reactivation stimulus in THP-1 cells. THP-1 cells were infected with TB40/E WT, ΔiP1, ΔiP2, or ΔiP(1 + 2) (MOI = 2) for 5 d and then treated with TPA to trigger viral reactivation. (A) RNA was isolated, and RT-qPCR was performed to monitor accumulation of iP1-, iP2-, and MIEP/dP-derived transcripts relative to H6PD. Data from 3 independent biological replicates (each performed in triplicate) are shown; SE is depicted. (B) Accumulation of viral proteins was measured during latency and following reactivation stimulus by immunoblotting. IE1 and IE2 (immediate early) and pUL44 and pUL135 (early) were measured. Tubulin was used as a loading control. A single experiment (representative of 3 independent experiments) is shown. (C) Total DNA was isolated at days 1 and 5 (latency) and at day 10 (reactivation). Viral genomes were quantified by qPCR using a primer pair specific to the noncoding β2.7 region of the HCMV genome relative to standard curve using the bacterial artificial chromosome clone of TB40/E. Viral genome copy number was then normalized to the cellular gene RNase P. Bars represent fold change over the number of viral genomes present at day 1 for each virus. Data from 3 independent biological replicates (each performed in duplicate) are shown; SE is depicted. Two-way ANOVA revealed that fold change in genome copy number is not statistically significant (ns) for any of the infection groups.
The Intronic Promoters Are Necessary for Efficient HCMV Reactivation.
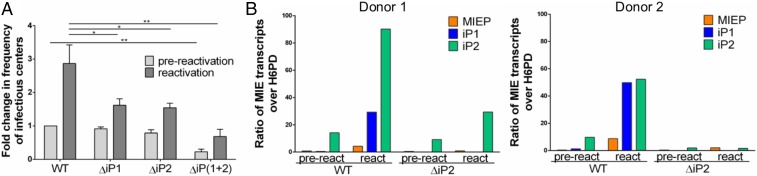
Primary CD34+ HPCs are considered the “gold standard” cell type for modeling HCMV latency and reactivation in vitro (21, 24–27). Moreover, CD34+ HPCs yield quantifiable numbers of progeny virus upon reactivation, which is a limitation of the THP-1 model. To evaluate whether iP1 and iP2 are required for reactivation in CD34+ HPCs, pure populations of infected (GFP+) CD34+ HPCs were isolated by cell sorting and seeded into long-term cultures over a stromal cell support to allow the establishment of latency (28). After 10 d, half of the culture was seeded by extreme limiting dilution onto fibroblasts in a cytokine-rich media to promote differentiation and reactivation. The other half of the culture was mechanically lysed to quantify the amount of infectious virus produced prior to the reactivation event. Reactivation in WT-infected cells produced a 3-fold greater frequency of infectious centers relative to its prereactivation control (Fig. 4A). By contrast, each of the ΔiP-mutant viruses exhibited a significant defect in reactivation, indicating a requirement for iPs for HCMV reactivation in primary CD34+ HPCs.
Fig. 4.
The intronic promoters are required for reactivation of HCMV from latency in CD34+ HPCs. CD34+ HPCs were infected with TB40/E WT, ΔiP1, ΔiP2, or ΔiP(1 + 2) expressing GFP as a marker for infection for 24 h (MOI = 2). Pure populations of infected (GFP+) CD34+ cells were isolated by FACS and maintained in long-term bone marrow culture for 10 d. (A) Viable CD34+ HPCs were seeded by limiting dilution onto monolayers of permissive fibroblasts in a cytokine-rich media to promote myeloid differentiation (reactivation, dark gray). An equivalent number of cells was mechanically lysed and seeded in parallel to determine the infectious virus present prior to reactivation (prereactivation, light gray). The frequency of infectious centers formed was determined 14 d later by ELDA from the fraction of GFP+ wells at each dilution. Data are expressed as fold change over the frequency of infectious centers produced by the WT virus prior to reactivation. Data from 3 independent biological replicates are shown; SE is depicted. Statistical significance was determined by multiple t tests comparing each mutant virus to the WT parental virus (*P value ≤ 0.05; **P value ≤ 0.005). (B) At day 10, RNA was collected from CD34+ HPCs latently infected with WT or ΔiP2 HCMV (prereactivation, pre). The remaining cells were plated in a modified “cell-free” reactivation assay absent coculture with permissive fibroblasts and in reactivation media enriched with 45 ng/mL of IL-6, G-CSF, and GM-CSF for 7 d before RNA was collected from adherent cells (react). RT-qPCR was performed to quantify discrete MIE transcripts relative to H6PD. Data from 2 independent biological replicates (qPCR reaction performed in triplicate) using cells from multiple donors are shown.
We next measured MIEP-, iP1-, and iP2-derived transcript accumulation in CD34+ HPCs from 2 donors infected with WT or ∆iP2 prior to (day 10 in culture) and following a reactivation stimulus (7 d post stimulus). The ΔiP2 recombinant was chosen because iP2-derived transcripts are the most abundant following TPA treatment in THP-1 cells. Consistent with the expression of these transcripts in THP-1 cells, iP-derived transcripts predominated following reactivation of WT virus compared to MIEP-derived transcripts (Fig. 4B). A global defect in induction of all MIE transcripts was observed for ΔiP2-infected cells when compared to WT-infected cells following the reactivation stimulus. As noted in THP-1 cells, iP2-derived transcripts were detected at a low level even during the latent period.
Discussion
The establishment of latency requires suppression, if not silencing, of the viral genes encoding the major immediate early transactivators, which drive viral gene expression and productive viral replication. A key unanswered question in HCMV biology concerns how viral gene expression is reinitiated following a reactivation stimulus. Collectively, our findings challenge the long-held paradigm that viral reactivation relies on de-repression of the MIEP to drive IE1 and IE2 expression and subsequent reentry into the virus lytic cycle. Rather, we find that the majority of UL123 and UL122 transcription during reactivation is driven by recently identified promoters within intron A of the MIE transcriptional locus (22). Disruption of these intronic promoters results in a striking defect in re-expression of viral genes and genome replication in the THP-1 and the primary CD34+ HPC latency model. These findings suggest that HCMV relies on multiple promoters to regulate MIE locus gene expression in a context-dependent manner. Promoter switching in the MIE locus during latency and reactivation may minimize spontaneous reactivation and limit the expression of viral antigens that may otherwise be detected by the immune system.
It has long been assumed that MIE gene expression that supports reactivation of the viral lytic cycle results from de-repression of the MIEP, and changes in transcription factor binding and chromatin have been detected at the MIEP upon reactivation (29–35). While our data do not preclude a role for the MIEP, iP-derived transcripts are the predominant drivers of viral IE gene re-expression following a reactivation stimulus (Fig. 2C), and iP-driven IE gene expression is required for efficient reactivation (Figs. 3 and 4). By contrast, MIEP-derived transcripts were detected at very low levels only at 12 and 14 dpi. It also remains a possibility that chromatin reorganization and transcription factor binding at the MIEP influences activity of the intronic promoters. Collectively, our data suggest that activation of the intronic promoters is the primary event (i.e., occurring before MIEP activation) required for HCMV reactivation.
The defect in reactivation resulting when one or both intronic promoter elements is deleted appears more pronounced in THP-1 cells than in primary CD34+ HPCs. These differences may be due to the heterogeneity of CD34+ primary cells compared to THP-1 cells, a homogenous population that can be triggered to reactivate in a more synchronous manner. Alternatively, these differences may also reflect the inherent limitations or technical differences of each model system. For example, the readout for reinitiation of viral gene expression in THP-1 cells is total IE protein in a pooled population of cells in which only 30 to 50% of the cells were initially infected, thus increasing the potential for very subtle changes to be masked. By contrast, the extreme limiting dilution assay used to quantify the number of latently infected CD34+ cells required to produce viral progeny is of greater sensitivity and is inherently subject to more donor variability. Although THP-1 cells do not produce robust titers of progeny viruses following reactivation stimulus (36), our work highlights the value of THP-1 cells as a model for transcriptional silencing and re-expression of HCMV gene expression during latency and reactivation.
The early burst of gene expression at 1 dpi (Fig. 1) and the noisy expression of transcripts from the iP2 element (Figs. 2C and 4B) is consistent with recent work in the field suggesting that viral transcription is not as silent during latency as previously presumed (37, 38). The initial burst of IE gene expression that we observed during the establishment of latency is likewise consistent with reports from other groups (39, 40). Collectively, the available data from both primary cell and cell line models indicate that viral gene expression is repressed, but not completely silenced during latency. More work is required to understand if early bursts of gene expression are important for the establishment of latency.
Notably, we consistently detected low levels of iP2-derived transcripts throughout latency (Figs. 2C and 4B), although IE proteins remained virtually undetectable (Fig. 1A and SI Appendix, Fig. S1). The sustained levels of iP2-derived transcripts in the absence of detectable levels of protein accumulation may suggest that the translation, as well as transcription, of MIE genes is regulated in a context-dependent manner (e.g., stress-induced translation), consistent with previously established correlates linking cellular stress to viral reactivation (41). For example, the discrete 5′ untranslated regions (UTRs) of the iP-derived transcripts contain multiple alternative AUG translation start sites which may be posited to suppress translational activity via leaky scanning under basal conditions while promoting translation during cell stress (42, 43). Such regulatory elements are common in viruses and appear to regulate translation in a similar manner (44–46). Furthermore, Arend et al. (47) have previously demonstrated that the 5′ UTR of MIE mRNAs plays a role in their translation and is required for efficient HCMV replication during lytic infection. In the case of iP-derived transcripts, such mechanisms may equip the virus with an additional point of control over IE gene expression, perhaps to minimize spontaneous viral reactivation.
In addition to expanding genomic coding potential in the human genome, alternative promoters play an important role in tailoring gene expression for various cell lineages, tissue types, and developmental stages (49). Furthermore, alternative promoter usage can produce multiple mRNAs encoding identical proteins, the synthesis of which is regulated by distinct 5′ UTRs. Herpesviruses also use alternative promoters to regulate viral gene expression. For example, Marek’s disease virus uses promoter switching to achieve differential expression of its immediate early transactivator ICP27 during lytic and latent infections (50). Furthermore, Kaposi’s sarcoma-associated herpesvirus encodes 6 splice variants of its ORF50 from 4 different promoters, resulting in multiple isoforms of the transactivator RTA, each with distinct viral transactivation potential (51). Epstein–Barr virus uses a promoter switching mechanism to maintain its distinct latency programs by driving differential expression of latency-associated genes (52–55). Potential mechanisms for promoter switching could include differences in the basal transcription machinery among different cell types, cell type-specific transcription factors, or epigenetic modifications to the viral genome. The significance of these promoter switching events to the outcome of infection remains to be fully defined. However, these studies and the present study suggest that promoter switching may play a prominent role in the lytic to latent switch during herpesvirus infections.
Understanding the regulation of alternative promoters may pave the way for the development of improved vaccines and gene therapy vectors. As deletion of the HCMV MIE intronic promoters limits potential for reactivation (Figs. 3 and 4) without impeding virus propagation in fibroblasts (SI Appendix, Fig. S3), cytomegalovirus-based vaccine vectors in which the minimal intronic promoter sequences are disrupted would be expected to limit reactivation while maintaining the potential to efficiently propagate the vector in replication-permissive fibroblasts. Additionally, inclusion of the intron A region of the MIE locus in gene therapy vectors driven by the HCMV promoter (MIEP) may enable efficacious gene expression in hematopoietic cells, a clinically important setting in which the core MIEP has minimal activity (12). While the cellular and/or viral factors that regulate HCMV intronic promoter activity during reactivation are currently unknown, our data suggest that defining such factors will be critical to understanding the molecular events controlling HCMV latency and reactivation.
Materials and Methods
THP-1 Latency Model.
THP-1 cells were infected as monocyte-like suspension cells at a density of 0.5 million cells per mL and a multiplicity of infection (MOI) of 2 (as determined by TCID50 using MRC-5 fibroblasts). The cell suspension was mixed by rocking every 30 min for 4 h, followed immediately by a “spinfection” at 450 × g for 20 min, yielding 30 to 50% infection (GFP+ by FACs) at 24 hpi. Cells were cultured for 5 dpi in nontissue culture-treated 6-well plates (1.5 million cells in 3 mL per well). During the 5-d latency period, infected cells continued to proliferate, and media was added to maintain a cell density of ≥1 million cells per mL. Viral genomes were maintained during the 5-d latency period (Fig. 3C). On day 5, cells from each experimental group were pooled and centrifuged at 120 × g for 7 min and then resuspended at 0.5 million cells per mL. Cells were treated with 100 nM TPA and plated on tissue culture-treated plates to promote monocyte-to-macrophage differentiation (and viral reactivation) or treated with DMSO as a solvent control and cultured nonadherently. Cells were washed in PBS at 24 h post TPA/DMSO treatment, and fresh media was added to the culture at 1 mL for every 0.5 million cells. TPA treatment resulted in differentiation of 50 to 75% of the treated cells; any cells that remained in suspension after 24 h of treatment were removed from the culture with 2 PBS washes, leaving only differentiated cells. Dead cells were removed from the plate with 2 PBS washes immediately prior to collection of proteins or nucleic acids.
Assays of Infectious Centers for Latency.
Frequency of reactivation for each mutant virus relative to WT in CD34+ HPCs was quantified as previously described (21, 26). Briefly, pure populations of CD34+ HPCs were infected with WT, ΔiP1, ΔiP2, or ΔiP(1 + 2) virus (MOI = 2) expressing GFP as a marker for infection. At 24 hpi, infected (GFP+) CD34+ cells were isolated by fluorescence-activated cell sorting (FACS) and incubated in long-term culture with a stromal cell support to maintain latency. At 10 dpi, viable CD34+ HPCs were seeded by limiting dilution onto monolayers of permissive MRC-5 fibroblasts (reactivation). An equivalent number of CD34+ HPCs were mechanically disrupted and seeded in parallel to quantify infectious virus present prior to reactivation (prereactivation). Frequency of infectious centers was quantified 14 d later by extreme limiting dilution analysis (ELDA) of GFP+ wells. Please see the SI Appendix for a detailed description of the Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jim Alwine and the laboratories of Dr. John Purdy and Dr. Anita Koshy at the University of Arizona for helpful discussion of the data presented in this manuscript. Mark Curry and the Arizona Cancer Center/Arizona Research Laboratories Division of Biotechnology Cytometry Core Facility provided expertise and assistance with flow cytometry. Research was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health Grants AI 079059 (F.G.) and AI 143191 (to F.G., N.J.M, and J.P.K.) and by a Pew Innovator Award (to F.G.). D.C.-M. is supported by a Postdoctoral Fellowship (18POST33960140) from the American Heart Association. J.C.B. is supported by a Postdoctoral Fellowship (129842-PF-16-212-01-TBE) from the American Cancer Society.
Footnotes
Conflict of interest statement: F.G., N.J.M., and J.P.K. have filed a patent for manipulation of intronic promoters to reduce reactivation of viral vaccines.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900783116/-/DCSupplemental.
References
- 1.Nogalski M. T., Collins-McMillen D., Yurochko A. D., “Overview of human cytomegalovirus pathogenesis” in Human Cytomegaloviruses: Methods & Protocols, Yurochko A. D., Miller W. E., Eds. (Humana Press, New York, 2014), chap. 2, pp. 15–28. [DOI] [PubMed] [Google Scholar]
- 2.Mocarski E. Jr, Shenk T., Griffiths P. D., Pass R., “Cytomegaloviruses” in Fields Virology, Knipe D. M., Howley P. M., Eds. (Lippincott Williams & Wilkins, Philadelphia, PA, 2013), vol. 2, pp. 1960–2014. [Google Scholar]
- 3.Stinski M. F., Meier J. L., “Immediate-early viral gene regulation and function” in Human Herpesviruses, Biology T., et al., Eds. (Cambridge University Press, Cambridge, UK, 2007), chap. 17. [Google Scholar]
- 4.Goodrum F., Human cytomegalovirus latency: Approaching the Gordian knot. Annu. Rev. Virol. 3, 333–357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins-McMillen D., Buehler J. C., Peppenelli M., Goodrum F., Molecular determinants and the regulation of human cytomegalovirus latency and reactivation. Viruses 10, E444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomsen D. R., Stenberg R. M., Goins W. F., Stinski M. F., Promoter-regulatory region of the major immediate early gene of human cytomegalovirus. Proc. Natl. Acad. Sci. U.S.A. 81, 659–663 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshart M., et al. , A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell 41, 521–530 (1985). [DOI] [PubMed] [Google Scholar]
- 8.Ghazal P., Lubon H., Fleckenstein B., Hennighausen L., Binding of transcription factors and creation of a large nucleoprotein complex on the human cytomegalovirus enhancer. Proc. Natl. Acad. Sci. U.S.A. 84, 3658–3662 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinski M. F., Roehr T. J., Activation of the major immediate early gene of human cytomegalovirus by cis-acting elements in the promoter-regulatory sequence and by virus-specific trans-acting components. J. Virol. 55, 431–441 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelbourn S. L., Kothari S. K., Sissons J. G., Sinclair J. H., Repression of human cytomegalovirus gene expression associated with a novel immediate early regulatory region binding factor. Nucleic Acids Res. 17, 9165–9171 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair J. H., Baillie J., Bryant L. A., Taylor-Wiedeman J. A., Sissons J. G., Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J. Gen. Virol. 73, 433–435 (1992). [DOI] [PubMed] [Google Scholar]
- 12.Stinski M. F., “Cytomegalovirus promoter for expression in mammalian cells” in Gene Expression Systems: Using Nature for the Art of Expression, Fernandez J. M., Hoeffler J. P., Eds. (Elsevier, Cambridge, MA, 1999), chap. 8, pp. 211–233. [Google Scholar]
- 13.Keller M. J., et al. , Reversal of human cytomegalovirus major immediate-early enhancer/promoter silencing in quiescently infected cells via the cyclic AMP signaling pathway. J. Virol. 81, 6669–6681 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan J., et al. , Breaking human cytomegalovirus major immediate-early gene silence by vasoactive intestinal peptide stimulation of the protein kinase A-CREB-TORC2 signaling cascade in human pluripotent embryonal NTera2 cells. J. Virol. 83, 6391–6403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X., et al. , Phorbol ester-induced human cytomegalovirus major immediate-early (MIE) enhancer activation through PKC-delta, CREB, and NF-kappaB desilences MIE gene expression in quiescently infected human pluripotent NTera2 cells. J. Virol. 84, 8495–8508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinshenker B. G., Wilton S., Rice G. P., Phorbol ester-induced differentiation permits productive human cytomegalovirus infection in a monocytic cell line. J Immunol 140, 1625–1631 (1988). [PubMed] [Google Scholar]
- 17.Arcangeletti M. C., et al. , Human cytomegalovirus reactivation from latency: Validation of a “switch” model in vitro. Virol. J. 13, 179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee C. H., et al. , Factors affecting human cytomegalovirus gene expression in human monocyte cell lines. Mol. Cells 9, 37–44 (1999). [PubMed] [Google Scholar]
- 19.Yee L. F., Lin P. L., Stinski M. F., Ectopic expression of HCMV IE72 and IE86 proteins is sufficient to induce early gene expression but not production of infectious virus in undifferentiated promonocytic THP-1 cells. Virology 363, 174–188 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Umashankar M., et al. , A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 7, e1002444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umashankar M., et al. , Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection. J. Virol. 88, 5987–6002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arend K. C., Ziehr B., Vincent H. A., Moorman N. J., Multiple transcripts encode full-length human cytomegalovirus IE1 and IE2 proteins during lytic infection. J. Virol. 90, 8855–8865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo K., Xu J., Mocarski E. S., Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. U.S.A. 93, 11137–11142 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodrum F. D., Jordan C. T., High K., Shenk T., Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: A model for latency. Proc. Natl. Acad. Sci. U.S.A. 99, 16255–16260 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrucelli A., Rak M., Grainger L., Goodrum F., Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J. Virol. 83, 5615–5629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caviness K., et al. , Complex interplay of the UL136 isoforms balances cytomegalovirus replication and latency. MBio 7, e01986 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buehler J., et al. , Opposing regulation of the EGF receptor: A molecular switch controlling cytomegalovirus latency and replication. PLoS Pathog. 12, e1005655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umashankar M., Goodrum F., Hematopoietic long-term culture (hLTC) for human cytomegalovirus latency and reactivation. Methods Mol. Biol. 1119, 99–112 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Reeves M. B., Lehner P. J., Sissons J. G., Sinclair J. H., An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 86, 2949–2954 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Reeves M. B., MacAry P. A., Lehner P. J., Sissons J. G., Sinclair J. H., Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U.S.A. 102, 4140–4145 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kew V. G., Yuan J., Meier J., Reeves M. B., Mitogen and stress activated kinases act co-operatively with CREB during the induction of human cytomegalovirus immediate-early gene expression from latency. PLoS Pathog. 10, e1004195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Connor C. M., Nukui M., Gurova K. V., Murphy E. A., Inhibition of the FACT complex reduces transcription from the human cytomegalovirus major immediate early promoter in models of lytic and latent replication. J. Virol. 90, 4249–4253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elder E., Sinclair J., HCMV latency: What regulates the regulators? Med. Microbiol. Immunol. 208, 431–438 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna B. A., Humby M. S., Miller W. E., O’Connor C. M., Human cytomegalovirus G protein-coupled receptor US28 promotes latency by attenuating c-fos. Proc. Natl. Acad. Sci. U.S.A. 116, 1755–1764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim E. T., et al. , SAMHD1 modulates early steps during human cytomegalovirus infection by limiting NF-kB activation. Cell Rep. 28, 434–448.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albright E. R., Kalejta R. F., Myeloblastic cell lines mimic some but not all aspects of human cytomegalovirus experimental latency defined in primary CD34+ cell populations. J. Virol. 87, 9802–9812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng S., et al. , Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. U.S.A. 114, E10586–E10595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shnayder M., et al. , Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio 9, e00013-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung A. K., Abendroth A., Cunningham A. L., Slobedman B., Viral gene expression during the establishment of human cytomegalovirus latent infection in myeloid progenitor cells. Blood 108, 3691–3699 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Taylor-Wiedeman J., Sissons P., Sinclair J., Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68, 1597–1604 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prösch S., Döcke W. D., Reinke P., Volk H. D., Krüger D. H., Human cytomegalovirus reactivation in bone-marrow-derived granulocyte/monocyte progenitor cells and mature monocytes. Intervirology 42, 308–313 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Calvo S. E., Pagliarini D. J., Mootha V. K., Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc. Natl. Acad. Sci. U.S.A. 106, 7507–7512 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma D. K., Bressler K., Patel H., Balasingam N., Thakor N., Role of eukaryotic initiation factors during cellular stress and cancer progression. J Nucleic Acids 2016, 8235121 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shabman R. S., et al. , An upstream open reading frame modulates Ebola virus polymerase translation and virus replication. PLoS Pathog. 9, e1003147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kronstad L. M., Brulois K. F., Jung J. U., Glaunsinger B. A., Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLoS Pathog. 9, e1003156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H. Y., Guan B. J., Su Y. P., Fan Y. H., Brian D. A., Reselection of a genomic upstream open reading frame in mouse hepatitis coronavirus 5′-untranslated-region mutants. J. Virol. 88, 846–858 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arend K. C., Lenarcic E. M., Moorman N. J., The 5′ untranslated region of the major immediate early mRNA is necessary for efficient human cytomegalovirus replication. J. Virol. 92, e02128-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balázs Z., et al. , Long-read sequencing of human cytomegalovirus transcriptome reveals RNA isoforms carrying distinct coding potentials. Sci. Rep. 7, 15989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davuluri R. V., Suzuki Y., Sugano S., Plass C., Huang T. H., The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 24, 167–177 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Strassheim S., et al. , Oncogenic Marek’s disease herpesvirus encodes an isoform of the conserved regulatory immediate early protein ICP27 generated by alternative promoter usage. J. Gen. Virol. 97, 2399–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 51.Wakeman B. S., Izumiya Y., Speck S. H., Identification of novel Kaposi’s sarcoma-associated herpesvirus Orf50 transcripts: Discovery of new RTA isoforms with variable transactivation potential. J. Virol. 91, e01434-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woisetschlaeger M., Yandava C. N., Furmanski L. A., Strominger J. L., Speck S. H., Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 87, 1725–1729 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaefer B. C., Strominger J. L., Speck S. H., Redefining the Epstein-Barr virus-encoded nuclear antigen EBNA-1 gene promoter and transcription initiation site in group I Burkitt lymphoma cell lines. Proc. Natl. Acad. Sci. U.S.A. 92, 10565–10569 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nonkwelo C., Skinner J., Bell A., Rickinson A., Sample J., Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J. Virol. 70, 623–627 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tempera I., Klichinsky M., Lieberman P. M., EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 7, e1002180 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.