Significance
The main stabilizer of the DNA double helix is not the base-pair hydrogen bonds but coin-pile stacking of base pairs, whose hydrophobic cohesion, requiring abundant water, indirectly makes the DNA interior dry so that hydrogen bonds can exert full recognition power. We report that certain semihydrophobic agents depress the stacking energy (measurable in single-molecule experiments), leading to transiently occurring holes in the base-pair stack (monitorable via binding of threading intercalators). Similar structures observed in DNA complexes with RecA and Rad51, and previous observations of spontaneous strand exchange catalyzed in semihydrophobic model systems, make us propose that some hydrophobic protein residues may have roles in catalyzing homologous recombination. We speculate that hydrophobic catalysis is a general phenomenon in DNA enzymes.
Keywords: DNA, hydrophobic catalysis, threading intercalation, DNA polymerase, RecA
Abstract
Hydrophobic base stacking is a major contributor to DNA double-helix stability. We report the discovery of specific unstacking effects in certain semihydrophobic environments. Water-miscible ethylene glycol ethers are found to modify structure, dynamics, and reactivity of DNA by mechanisms possibly related to a biologically relevant hydrophobic catalysis. Spectroscopic data and optical tweezers experiments show that base-stacking energies are reduced while base-pair hydrogen bonds are strengthened. We propose that a modulated chemical potential of water can promote “longitudinal breathing” and the formation of unstacked holes while base unpairing is suppressed. Flow linear dichroism in 20% diglyme indicates a 20 to 30% decrease in persistence length of DNA, supported by an increased flexibility in single-molecule nanochannel experiments in poly(ethylene glycol). A limited (3 to 6%) hyperchromicity but unaffected circular dichroism is consistent with transient unstacking events while maintaining an overall average B-DNA conformation. Further information about unstacking dynamics is obtained from the binding kinetics of large thread-intercalating ruthenium complexes, indicating that the hydrophobic effect provides a 10 to 100 times increased DNA unstacking frequency and an “open hole” population on the order of 10−2 compared to 10−4 in normal aqueous solution. Spontaneous DNA strand exchange catalyzed by poly(ethylene glycol) makes us propose that hydrophobic residues in the L2 loop of recombination enzymes RecA and Rad51 may assist gene recombination via modulation of water activity near the DNA helix by hydrophobic interactions, in the manner described here. We speculate that such hydrophobic interactions may have catalytic roles also in other biological contexts, such as in polymerases.
The forces that stabilize the DNA double helix are a prerequisite for the secure storage of genetic information but their modest strength is also necessary for the efficient processes of replication, transcription, recombination, and repair systems—wherein thermal fluctuations, or “breathing,” play an important role (1, 2). A generally shallow free-energy landscape implies that the average structures and dynamics of DNA and RNA systems are easily perturbed and shifted by even weak-binding agents and altered solvent conditions. Indeed, the response to bending perturbation is an attribute that nature can use for recognition purposes (3–5). However, small molecules and ions also are crucially important, either as structurally defined elements (water bridges, magnesium or spermidine ions at specific positions in condensed DNA, etc.) or for their effect on the dielectric medium around the molecule, with less-well-defined interaction structures.
In the current context of destabilizing double-stranded DNA we wish to refer to 3 seminal contributions made more than 60 y ago—apart from the discovery by Crick and Watson of the double-helix structure: 1) results from Rosalind Franklin showing that the conformation of DNA is highly sensitive to hydration (6), 2) Herskovits demonstrating (7), as confirmed by others (8–11), that the DNA double helix is stabilized mainly by hydrophobic and dispersive stacking interactions and less by the hydrogen bonding between matched bases that most textbooks still refer to, and 3) the discovery of “breathing fluctuations” of DNA (12–14) paving the way for today’s single-molecule studies on how the regulatory machinery gets access to the “interior” of the double helix for reading and manipulating the genome (1).
A special class of fluctuations results in extension of the DNA duplex along its double-helical axis, causing the faces of adjacent bases to transiently unstack from one another without breaking the base pairs, creating holes in the hydrophobic helix interior. Such fluctuations, which we denote as “longitudinal breathing,” should be involved in transition states leading to intercalation of molecules between the bases and base flipping (e.g., B-to-Z transition), but also to metastable conformations such as the recently reported overstretched but still double-stranded “Σ-DNA” (15, 16); the original suggestion that such a structure indeed exists was made much earlier (17). In systematic thermochemical experiments Kool and coworkers (18, 19) deduced that the unstacking energy of nucleobases is 1.1 to 2.0 kcal/mol in buffer solution. This energy range compares well with the 1.6 kcal/mol activation energy of Σ-DNA (15, 16). The theoretical description of stacking association is challenging since the net base-stacking forces act in a complex and context-specific manner, the stacking forces being balanced with many other energy contributions (20). Anisotropy of dispersion interactions together with the hydrophobic stabilization and steric repulsion contribute to the parallel coin-pile stacking which determines both characteristics such as the stiffness of the double helix (a large persistence length) as well as various chemical consequences of unstacking dynamics.
We report here the surprising finding that water-miscible ethylene glycol ethers, such as PEG-400 and diglyme, affect double-stranded DNA in a way that differs significantly from other common organic solvents where denaturation, compaction, or aggregation are frequently observed (21). By contrast we find that ethylene glycol ether mixtures (up to 50% by weight) reduce the base-stacking energy and promote longitudinal breathing. A softening of the nucleobase stacking is visible as a significant decrease of the persistence length, without compromising the overall B conformation or the integrity of base pairing. As a result of thermal unstacking fluctuations, holes in the DNA base stack appear, which can be detected by adding large metal-organic compounds that thread themselves to reach an intercalative binding geometry. The binding kinetics of such threading intercalation into DNA is strongly accelerated by the presence of the hydrophobic agents. In single-molecule optical tweezers experiments the overstretching force of single DNA molecules is found to be reduced about 30% by these agents. In addition, a hyperchromicity of 3 to 6% suggests that the average interplanar distance between the stacked base pairs is slightly increased.
We suggest that longitudinal breathing may be involved in homologous gene recombination where RecA and Rad51 proteins mediate strand exchange reaction in prokaryotes and eukaryotes, respectively. These proteins disrupt the base stacking every 3 bases and elongate the DNA (22, 23). Despite the importance of recombinases in biological and medicinal context (cancer, gene therapy, sterility, etc.), and several decades of intense research, the mechanisms involved in performing a search for homology and executing strand exchange are not yet understood at an atomistic level (24). Recombinases must simultaneously both soften the DNA duplex, which is mainly stabilized through base stacking, as well as increase the fidelity of sequence recognition, which can only be achieved by strengthening the hydrogen bonds involved in base pairing. We earlier provided some evidence that a model system of aqueous polyethylene glycol (PEG) can achieve these 2 seemingly conflicting objectives (25, 26) and we here demonstrate that ethylene glycol ethers, in addition to promoting base unstacking, also strengthen base pairing, thereby increasing the energy difference between matched and mismatched sequences.
Our observation of base unstacking in hydrophobic or semihydrophilic water mixtures could have great general impact, hydrophobic catalysis potentially having a role in enzyme function, in particular of recombinases and DNA polymerases. Despite numerous studies of DNA in various solvents, specific longitudinal unstacking in a hydrophobic environment has to our best knowledge never been reported before.
Results
As shown in SI Appendix, Fig. S1, neither 40% PEG nor 20% diglyme significantly changes the circular dichroism (CD) spectrum of DNA: The overall B-DNA conformation is unaffected and any transformation into Ψ-DNA or other exotic conformations can be excluded. SI Appendix, Fig. S2 shows that adding PEG or diglyme to DNA in pure buffer causes a slight (3 to 6%) hyperchromicity of DNA at the usual 260-nm absorbance peak which can be explained by some degree of unstacking of nucleobases. Upon adding more buffer to dilute the ethylene glycol ether concentration, the hyperchromicity vanishes.
Glyoxal reaction experiments (selective for single strands; see SI Appendix, section 1c and Figs. S11 and S12) indicate no increase in frequency of unpaired bases in PEG and diglyme versus pure buffer, but rather the opposite. While absorbance spectra show that 60 mM glyoxal quickly denatures ctDNA in pure buffer at 50 °C, there is no significant reaction in 20 or 40% PEG. The same outcome was noticed with 10 and 20% diglyme and up to 20 mM glyoxal.
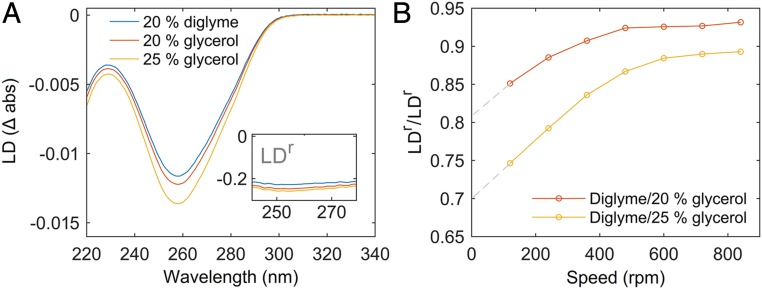
Flow linear dichroism (LD) spectra (for an explanation of LD see SI Appendix, section 1d) of DNA in diglyme and glycerol at varying shear rates are presented in Fig. 1 and SI Appendix, Figs. S13–S20. The measured viscosity of 20% diglyme (1.84 mPa·s) is between those measured for 20 and 25% glycerol (1.64 and 1.96 mPa·s, respectively). A range of glycerol concentrations was chosen to cover the viscosity for 20% diglyme with certainty. Fig. 1B shows the ratio between reduced LD (LDr = LD/Aiso) values in diglyme and glycerol at different shear gradients. The LD results provide 2 pieces of information: the average base orientation and the persistence length (SI Appendix, section 1d). The constant LDr as a function of wavelength over the 260-nm absorption band indicates that the bases are approximately perpendicular to the DNA helix axis, like in B-DNA. LDr is also directly proportional to the ratio P/L, of the persistence length over the contour length (27), which can then be compared between diglyme and glycerol solutions of the same viscosity. When extrapolated to zero shear rate, where DNA adopts its native relaxed configuration, the LDr of DNA in 20% diglyme is found to be between 0.70 and 0.81 of that in glycerol. This value translates to a persistence length of 35 to 41 nm, taking 50 nm as a reasonable value in up to 25% glycerol where B-DNA is unperturbed (28, 29).
Fig. 1.
Flow LD confirms B-DNA and indicates shorter persistence length in diglyme. (A) DNA LD spectra (720 rpm; see SI Appendix, Figs. S14–S16 for all speeds) with wavelength-independent LDr is consistent with bases near perpendicular to helix axis in diglyme and glycerol. (B) LDr in diglyme is less negative than in glycerol of the same viscosity. Hence, DNA persistence length is shortened in diglyme. Extrapolation to zero flow gradient indicates persistence length in diglyme between 70 and 81% of that in glycerol (see text and SI Appendix, section 1d).
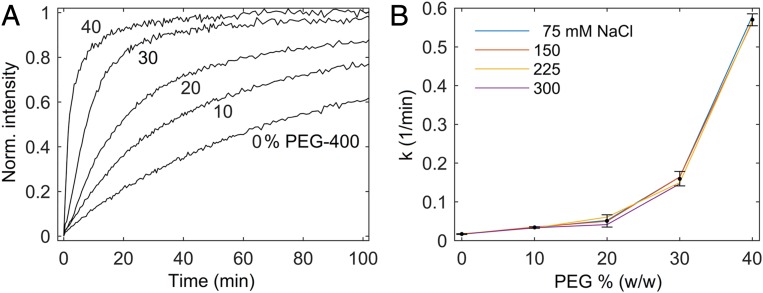
Threading intercalation (SI Appendix, section 1b) of [μ‐bidppz(phen)4Ru2]4+ into DNA (structure and depiction of intercalation in SI Appendix, Fig. S5) is dramatically accelerated by the presence of PEG, as demonstrated in Fig. 2. Here we report PEG-400 and diglyme (SI Appendix, Fig. S6) but the effect appears general with ethylene glycol ethers including cyclic ones such as 1,4-dioxane (SI Appendix, Fig. S7). By contrast no catalytic effects are observed with poly-alcohols like glycerol, poly(vinyl alcohol) (PVA), or dextran (SI Appendix, Figs. S8–S10). The kinetic traces are excellently fit to a biexponential model (Fig. 2B; details in SI Appendix, section 4). The major, fast exponential can be assigned to the true threading intercalation step, while a minor, slower exponential is probably associated with a rearrangement of the intercalated species (30, 31). SI Appendix, Table S1 presents DNA melting temperatures (Tm) for the conditions in Fig. 2B. As expected, a lower Tm is observed in PEG solutions. If faster intercalation were simply due to partial melting of DNA, an increased Tm upon adding salt would suppress intercalation. As seen in Fig. 2B, the rate constant appears salt-independent.
Fig. 2.
Threading intercalation accelerated by PEG. The complex [μ‐bidppz(phen)4Ru2]4+ (structure in SI Appendix, Fig. S5) is only fluorescent when intercalated in DNA. (A) Representative fluorescence kinetic traces of intercalation in PEG. (B) The major exponential rate constant k (fitting in SI Appendix, section 3) is independent of salt concentration. Salt concentration pooled to estimate SD (error bars ±2 SD, data missing for 40%, 300 mM due to precipitation).
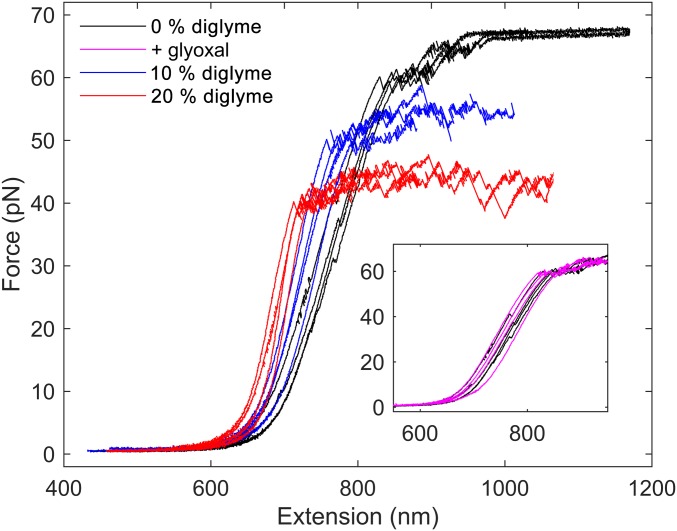
The results from single-molecule DNA pulling experiments using optical tweezers are presented in Fig. 3 in the presence and absence of diglyme. While contour length L and persistence length P do not change significantly according to the pulling experiment analysis, the overstretching force is dramatically reduced from about 60 pN to 40 pN while the stretch modulus S increases from about 350 pN to 650 pN upon addition of 20% diglyme (SI Appendix, Table S2). Pulling experiments on DNA preincubated with glyoxal and diglyme were also performed and, as expected from the glyoxal reaction measurements above, no difference from native DNA was found (Fig. 3, Inset). Thus, DNA stability is decreased by diglyme in agreement with melting experiments in PEG (SI Appendix, Table S1). However, since the 2 main contributions to total DNA stability of relevance are stacking and base pairing, and the latter is apparently rather increased by a hydrophobic environment (S is larger), the conclusion is that a weakened stacking stability has lowered the overstretching force.
Fig. 3.
Optical tweezers force spectroscopy. DNA stability measured by overstretch force is significantly weakened in 20% diglyme. (Inset) Absence of glyoxal reaction in 20% diglyme proves that loss of DNA stretch stability is not due to local denaturation or increased unpairing breathing. Increased slope of force-elongation curve in the main figure is consistent with decreased unpairing breathing in diglyme.
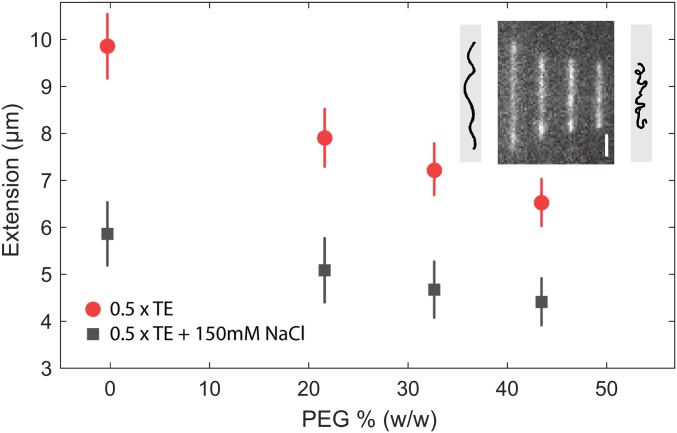
To further investigate the physical properties of DNA at the single-DNA-molecule level we used nanofluidic channels (32, 33). These measurements complement the optical tweezers experiment since they probe the physical properties of double-stranded DNA when it is not extended to its full contour length, a regime that is not easily analyzed with optical tweezers. Measurements were done for λ-DNA stained with YOYO-1 in buffer with and without 150 mM NaCl added, respectively, at increasing concentrations of PEG-400 (up to 40% wt/wt) in nanochannels with dimensions 100 × 150 nm2. At both conditions the DNA extension along the channels decreases with increasing PEG-400 concentration, and the effect is more pronounced at the lower ionic strength (Fig. 4). This demonstrates that PEG-400 softens the bending rigidity of double-stranded DNA (dsDNA) significantly.
Fig. 4.
Nanochannel experiments. Extension of λ-DNA with and without 150 mM NaCl versus PEG concentration. DNA confined in 100 × 150 nm2 channels. (Inset) A montage of fluorescence images of DNA molecules corresponding to the 4 red points. (Scale bar, 2 µm.) Cartoons show how reduced persistence length also reduces extension.
Discussion
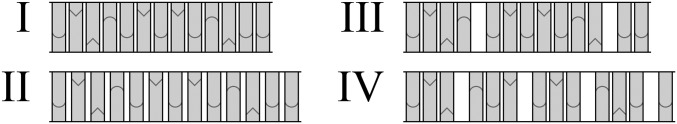
We present several pieces of evidence which coherently support the hypothesis that semihydrophobic water-miscible agents, such as ethylene glycol ethers, affect structure and dynamics of DNA in a way that facilitates interactions that depend on the stacking stability of the double helix rather than the hydrogen-bond pairing stability. We denote the effect “hydrophobic catalysis” and the unstacking dynamics as “longitudinal breathing.” Molecular crowding effects, sometimes associated with PEG, are shown to be insignificant in the current context (discussed below). Our results may be discussed in terms of simplistic models for DNA deformation; in a sketch (Fig. 5) 3 different distortions of B form DNA (I) are envisaged: a homogeneously slightly stretched and unwound structure (II), an inhomogenously unstacked DNA (III), and a repetitive inhomogeneous conformation (IV), the latter similar to the DNA structure found in RecA and Rad51 complexes (22, 23, 34, 35).
Fig. 5.
Simplistic models of DNA unstacking: (I) normal B form, (II) slightly extended and unwound B-DNA, (III) inhomogeneously extended B-DNA with holes, and (IV) inhomogeneously extended B-DNA with repeating base triplets and holes [cf. Σ DNA (16)].
Threading Intercalation as a Probe for Hole Formation.
The bulky dumbbell-shaped ruthenium complexes are known to bind extremely slowly (hours/days at room temperature) to DNA by threading one of the Ru moieties through the stack of bases, to end up in an equilibrium geometry with one Ru2+ ion in each groove (36). The strongly accelerated threading intercalation rate observed in Fig. 2 can be related to another case of hydrophobic catalysis, namely where PEG accelerates the rate of spontaneous strand exchange in DNA (26, 37). For purely steric reasons, holes of significant size (0.3 to 0.5 nm) must open up in the DNA base stack to allow the voluminous ruthenium complex to thread itself through. From earlier DNA pulling experiments on the same complex, an estimate of a 0.33-nm hole size was made (38). One may envisage that this distortion could occur either by classical breathing (unpairing), by longitudinal breathing (unstacking), or as a combination of both.
Destabilized Base Pairing Can Be Excluded.
The salt-compensated DNA melting curves and kinetic traces, and absence of any glyoxal reaction, indicate that the destabilization of DNA that we observe in the presence of ethylene glycol ethers is not due to destabilized base pairing. DNA melting temperatures (SI Appendix, Table S1; salt concentrations as in Fig. 2B) confirm that PEG indeed lowers the Tm of DNA, but that the intercalation rate constant is independent of salt concentration. For example, both Tm and k are larger in 30% PEG with 300 mM NaCl than in pure buffer and 75 mM NaCl.
The glyoxal experiments are important in this context as they indicate no increase in frequency of unpaired bases in PEG and diglyme, although PEG is superior to diglyme in protecting DNA from glyoxal. Absence of unpaired bases in the presence of PEG and diglyme indicates that interstrand breathing is almost completely suppressed. This result is not entirely unexpected, since a hydrophobic environment will strengthen hydrogen bonds by decreasing the amount of water molecules that can act as competing hydrogen bonding partners. Stronger DNA base pairing has been noticed in nonpolar lipid contexts (39).
DNA Conformation and Stability.
CD and LD spectra of DNA in diglyme and PEG (Fig. 1A and SI Appendix, Figs. S1, S14, S17, and S20) clearly exclude significant conformational alterations from the B form. The differential and normalized nature of LDr makes this technique particularly sensitive to changes in the spatial orientation of transition moments in the DNA bases. Even a subtle overall base tilt will cause a significant slope in LDr(λ) over the 260-nm band, as observed for A DNA in 20% ethanol or at low relative humidity in a PVA matrix (40–42).
The single-molecule DNA pulling experiments (Fig. 3) show that presence of diglyme dramatically reduces cohesive stacking forces: The overstretching force decreased from 60 pN to 40 pN while the stretch modulus increased from about 350 pN to 650 pN upon addition of 20% diglyme (SI Appendix, Table S2). Experiments on DNA preincubated with glyoxal and diglyme showed no difference from native DNA, evidencing the absence of unpaired bases.
The hypothetical models II, III, and IV all assume that the decreased cohesive stacking forces lead to longitudinal breathing fluctuations, resulting in smaller or larger gaps between the bases. All 3 models have support from the hyperchromicity results, as well as from the flow LD and nanochannel experiments, indicating increased flexibility of DNA, while only III and IV are compatible with the accelerated threading intercalation results.
The extension of DNA in nanochannels, as shown in Fig. 4, decreases with increasing PEG concentration—consistent with an increased flexibility (shorter persistence length; see Fig. 4, Inset) However, the apparent extension also depends on several other parameters, such as the effective channel width and any interactions with the walls. This makes it difficult to directly relate the results to the decrease in persistence length concluded from the flow-LD measurements, but the experiments are clearly in qualitative agreement with each other.
Frequency of Hole Formation.
One may estimate a minimum hole opening frequency from the threading intercalation kinetics. Assume that 1 complex intercalated corresponds to the creation of 1 hole. Let N holes appear over time T. The probability of finding the first hole during Δt is Δt/T. The exponential rate constant k in in terms of probability is
which simplifies to k = N/T. The experimental k values in 40% PEG (∼0.57 min−1) roughly indicate that 1 hole appears every 1.75 min between any 2 stacking bases.
An alternative estimate is based on the relation between hyperchromicity and base separation (43, 44) (SI Appendix, section 8). The observed 3.5% hyperchromicity of DNA in 20% PEG/diglyme corresponds to an average base separation increase of 3% (i.e., from 0.345 to 0.355 Å). As shown in SI Appendix, this may be interpreted in terms of an inhomogeneous distribution, with about 4 full holes per 100 base pairs. However, there is also the possibility, in the limiting case of a completely homogeneous distribution, that hole formation is due to more rare thermal fluctuations and that increase in hyperchromicity and decrease in persistence length are due to incremental softening of all base–base contacts. Even a quite small decrease in steric stacking repulsion should lead to a significant decrease in persistence length P, in agreement with our experiments. In contrast to the electrostatic effects (phosphate–phosphate repulsion) which depend on ion environment and dielectric medium effects, the “steric persistence” is mainly determined by coin-pile repulsion with a r−12 dependence. From statistical mechanical work by Schellman (45) and Wilson and Schellman (46) the persistence length P may be related to the local bend angle θ between adjacent bases. Approximate relations of P to base-pair thickness lB and bend angle θ from one base to the next (assuming a hinge-like bending mechanism) can be applied to get a feeling for the decreased stiffness of the DNA double helix:
where Δ2 is the SD of angle θ. With P = 50 nm and lB = 0.345 nm we have from that θ = 6.73° (SD 0.791). A reduction of P with 10% to P = 45 nm thus, with lB = 0.355 nm (as deduced above), corresponds to θ = 7.20° (SD 0.904). In terms of the schematic models in Fig. 5, we may consider the “ground state” B-DNA (I) to have an angular “wiggle” of the base-plane normal relative to an average helix axis that increases slightly (average 0.47°) when passing to the “solvent-excited” DNA (II). The effect of large thermal fluctuations (III) is what we probe by the threading intercalation scenario. Finally, the fourth hypothetical structure (IV) represents the DNA structure observed in complex with RecA and Rad51 (16, 22, 23, 34). It is a strongly distorted DNA structure (50% elongated compared to B-DNA) and we have no indication that it should occur in the current hydrophobic environments but be formed under rather more extreme pulling conditions or in the presence of potentially intercalating protein residues (15, 16, 22, 23). Without proteins contributing precise interactions, structure IV would be discouraged by entropic considerations. We conclude that II and III are both very likely representations of the current solvent effects and may coexist.
Molecular Crowding?
The catalytic effect we observe is not an excluded volume (“molecular crowding”) or osmotic effect (47, 48). Dextran, a typical crowding agent and significantly more hydrophilic than PEG, we found has no apparent accelerating effect on the intercalation rate. Furthermore, the average molecular size of PEG-400 is too small to exert any significant volume of exclusion, and diglyme and dioxane are even smaller. Another argument against molecular crowding is our observation that in nanofluidic channels the apparent length of DNA is shortened by presence of PEG, in contrast to the elongation previously observed when crowding is significant (49).
Fundamental studies by Sugimoto and coworkers (50, 51) early showed how PEG can affect the thermodynamic stability of DNA. In addition to what is generally referred to as molecular crowding effects, correlations to water activity were concluded along the lines of earlier work by Spink and Chaires (52). However, the picture is complex as water activity is affected via several parallel mechanisms such as electrostatic, solvent viscosity, and dehydration effects—in combination with the excluded volume and osmotic effects (53, 54). In our case dehydration is judged less important since dioxane, diglyme, and PEG give very similar effects despite their widely different hydration properties. Hydrophobic effects thus seem to stand out as rather special in our context.
DNA Conformation in Other Solvents.
A variety of conformations were early characterized in humid DNA fibers by X-ray diffraction (6, 55) and identified by LD in humid stretched PVA matrix and other solvents (41, 42). The A form is found in PVA at 75% relative humidity and in 25% ethanol water mixtures. It shows negative but sloping LDr (=LD/A) as function of wavelength due to tilt of base pairs while the B form displays a flat, almost constant feature, the bases being approximately perpendicular to the helix axis. Similarly, in ethanolic solvent B and A forms are found at low and high ethanol concentrations, respectively. LD spectra agree reasonably well with Arnott’s structures applying exciton theory to the coupling of the pi-electron transitions in the base plane (40–42). The conformational variations have been linked theoretically to hydration patterns (56, 57). A variation found in methanol–ethanol–buffer mixtures (58–60) is a fully denatured form as judged from hyperchromicity and CD. A weak positive flow LD indicates only slightly ordered bases with planes more parallel than perpendicular to the flow direction and speculated to point outward, dwelling in the nonpolar solvent. The native B form is immediately restored if adding water or intercalating drugs, showing that the 2 DNA strands are never separated far from each other. Further polymorphs of DNA have been described in the literature (for a catalog with CD spectra, see ref. 61). A useful comparison has been made to how much different solvents unwind DNA (62). In no case, however, do we identify any features suggesting that we might not have B-form DNA in our ethylene glycol ether mixtures.
To understand the behavior of DNA in a hydrophobic or semihydrophobic environment we should note that we are not dealing with a typical “poor” solvent. PEG-400 and diglyme are completely miscible with water and from LD spectra we can exclude that any A-like conformation is adopted. An atomic force microscopy experiment (63) shows that pulling double-stranded DNA into a poor solvent (1-propanol or diethylbenzene) leads to complete dissociation into single strands (64). The authors speculate whether unwinding/splitting of dsDNA in a low-polarity microenvironment may be exploited by helicases. PEG is not known as a particularly hydrophobic species but can readily bind substantial amounts of water. However, at high concentrations the rigid DNA and the flexible PEG are, according to Flory’s theory, immiscible and DNA collapses above a critical concentration (65).
Concluding Remarks
From what has been learned here, we arrive at a picture of DNA in aqueous ethylene glycol ether mixtures that retains a B-like conformation which is base-paired but has a softer coin-pile stacking of the bases. Thermal fluctuations can be anticipated to more easily favor transient unstacking, which we denote as longitudinal breathing. All experimental results support our hypothesis that the ethylene glycol ethers create holes in the DNA helix by promoting base unstacking, which provides a hydrophobic mechanism for catalysis of intercalation.
From different pieces of evidence, we have made estimates of how much the presence of ethylene glycol ethers promotes the generation of unstacked holes. While CD is somewhat less sensitive to a slight unwinding and unstacking, the hyperchromicity increases rapidly with increasing base separation and could be used for estimates of longitudinal breathing opening dynamics corresponding to an average base separation, typically increasing by 3% (discussed above). Such a small difference is hardly visible as an increased contour length, but a decrease in steric stacking repulsion should lead to a significant decrease in persistence length as is indeed observed here.
One may envisage both the occurrence of larger fluctuations leading to large openings (“holes”) explaining the threading intercalation catalysis, as well as more homogeneous subtle liberations of the base stacking, so that the observed decrease of persistence length be explained by slightly widened distribution of angles between base planes. We cannot narrow down the mechanism to either of these scenarios but assume that both are present in determining the dynamics of DNA. A take-home message appears to be that hydrophobic catalysis results in a decrease of the hydrophobic stacking energy. That our ethylene glycol ethers reduce the hydrophobic stacking energy because of a modulated chemical potential of water is an attractive model with interesting consequences: it is expected to lead to openings of slots—like what is postulated in Σ-DNA (15, 16) and documented by crystallography in DNA complex with RecA and Rad51 (22, 23). We deliberately leave the question of exact mechanism of hydrophobic catalysis open: Unspecific solvent effects may combine with more specific hydrophobic interactions of the DNA with the added ethylene glycol ethers which may generally adopt amphiphilic conformations. We note that absence of hydrophobic catalysis with dextran and other hydrophilic alcohols indicates that molecular crowding (excluded volume osmosis) is not a significant effect.
We propose that hydrophobic catalysis may be exploited somehow by RecA and Rad51, and possibly also other DNA-operating enzymes. It has not escaped our notice that the L2 loop in RecA, containing several hydrophobic residues, could modulate the chemical potential of water near the DNA in analogy with what has been observed in this study. It would be interesting also to search for hydrophobic effects in interactions of DNA regulatory proteins where mechanistically important local motions in the ∼100-μs range have been detected in single-molecule LD experiments (66).
Materials and Methods
DNA conformation was studied using polarized (CD and LD) and conventional absorbance spectroscopy. The light-switching (fluorescent when intercalated) thread-intercalating ΔΔ-[μ‐bidppz(phen)4Ru2]4+ was used to probe for unstacked holes between bases. Glyoxal was used to exclude increased unpairing breathing in the presence of cosolutes. Single DNA molecules were observed using optical tweezers and nanochannels, the first method quantifying the decrease in B-DNA stability and the second method providing direct observation of decreased persistence length. Thorough explanations of materials and methods used are provided in SI Appendix, section 1.
Supplementary Material
Acknowledgments
This work was supported by Swedish Research Council Grant 2015-04020 (to B.N.); Swedish Research Council Grant 2015–5062 and Olle Engqvist Foundation Grant 2016/84 (to F.W.); NIH Grant R01-HG006851 (to K.D.D.); and NIH Grant R01GM032543 and US Department of Energy Office of Basic Energy Sciences Nanomachine Program Contract DE-AC02-05CH11231 (to C.J.B.). We thank Irfan Shaukat for early work (2010).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909122116/-/DCSupplemental.
References
- 1.von Hippel P. H., Johnson N. P., Marcus A. H., Fifty years of DNA “breathing”: Reflections on old and new approaches. Biopolymers 99, 923–954 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak C. H., Unraveling base stacking driving forces in DNA. J. Phys. Chem. B 120, 6010–6020 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Pasi M., et al. , μABC: A systematic microsecond molecular dynamics study of tetranucleotide sequence effects in B-DNA. Nucleic Acids Res. 42, 12272–12283 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rossetti G., et al. , The structural impact of DNA mismatches. Nucleic Acids Res. 43, 4309–4321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reymer A., Zakrzewska K., Lavery R., Sequence-dependent response of DNA to torsional stress: A potential biological regulation mechanism. Nucleic Acids Res. 46, 1684–1694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin R. E., Gosling R. G., Molecular configuration in sodium thymonucleate. Nature 171, 740–741 (1953). [DOI] [PubMed] [Google Scholar]
- 7.Herskovits T. T., Nonaqueous solutions of DNA: Factors determining the stability of the helical configuration in solution. Arch. Biochem. Biophys. 97, 474–484 (1962). [Google Scholar]
- 8.Sínanoĝlu O., Abdulnur S., Hydrophobic stacking of bases and the solvent denaturation of DNA. Photochem. Photobiol. 3, 333–342 (1964). [Google Scholar]
- 9.Frank-Kamenetskii M., DNA chemistry. How the double helix breathes. Nature 328, 17–18 (1987). [DOI] [PubMed] [Google Scholar]
- 10.Frank-Kamenetskii M. D., Biophysics of the DNA molecule. Phys. Rep. 288, 13–60 (1997). [Google Scholar]
- 11.Yakovchuk P., Protozanova E., Frank-Kamenetskii M. D., Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 34, 564–574 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Englander S. W., A hydrogen exchange method using tritium and sephadex: Its application to ribonuclease. Biochemistry 2, 798–807 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Printz M. P., Vonhippel P. H., Hydrogen exchange studies of DNA structure. Proc. Natl. Acad. Sci. U.S.A. 53, 363–370 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Von Hippel P. H., Printz M. P., Dynamic aspects of DNA structure as studies by hydrogen exchange. Fed. Proc. 24, 1458–1465 (1965). [PubMed] [Google Scholar]
- 15.Bosaeus N., et al. , Tension induces a base-paired overstretched DNA conformation. Proc. Natl. Acad. Sci. U.S.A. 109, 15179–15184 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosaeus N., et al. , A stretched conformation of DNA with a biological role? Q. Rev. Biophys. 50, e11 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Smith S. B., Cui Y., Bustamante C., Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–799 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Kool E. T., Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev.Biophys. Biomol. Struct. 30, 1–22 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Gao J., Liu H., Kool E. T., Expanded-size bases in naturally sized DNA: Evaluation of steric effects in Watson-Crick pairing. J. Am. Chem. Soc. 126, 11826–11831 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Šponer J., et al. , Nature and magnitude of aromatic base stacking in DNA and RNA: Quantum chemistry, molecular mechanics, and experiment. Biopolymers 99, 978–988 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Lowe M. J., Schellman J. A., Solvent effects on dinucleotide conformation. J. Mol. Biol. 65, 91–109 (1972). [DOI] [PubMed] [Google Scholar]
- 22.Chen Z., Yang H., Pavletich N. P., Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–4 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Xu J., et al. , Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat. Struct. Mol. Biol. 24, 40–46 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentiss M., Prévost C., Danilowicz C., Structure/function relationships in RecA protein-mediated homology recognition and strand exchange. Crit. Rev. Biochem. Mol. Biol. 50, 453–476 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Feng B., “DNA strand exchange and hydrophobic interactions between biomolecules,” PhD thesis, Chalmers University of Technology, Gothenburg, Sweden (2015).
- 26.Feng B., Westerlund F., Nordén B., Evidence for hydrophobic catalysis of DNA strand exchange. Chem. Commun. (Camb.) 51, 7390–7392 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Nordén B., Kubista M., Kurucsev T., Linear dichroism spectroscopy of nucleic acids. Q. Rev. Biophys. 25, 51–170 (1992). [DOI] [PubMed] [Google Scholar]
- 28.Hofrichter J. S., Schellman J. A., “The optical properties of oriented biopolymers” in The Jerusalem Symposia on Quantum Chemistry and Biochemistry, Bergmann E. D, Pullman B, Eds. (Jerusalem Academy of Sciences and Humanities, Jerusalem, 1973). [Google Scholar]
- 29.Nordén B., Seth S., Structure of strand-separated DNA in different environments studied by linear dichroism. Biopolymers 18, 2323–2339 (1979). [DOI] [PubMed] [Google Scholar]
- 30.Nordell P., et al. , DNA polymorphism as an origin of adenine-thymine tract length-dependent threading intercalation rate. J. Am. Chem. Soc. 130, 14651–14658 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Westerlund F., Wilhelmsson L. M., Nordén B., Lincoln P., Monitoring the DNA binding kinetics of a binuclear ruthenium complex by energy transfer: Evidence for slow shuffling. J. Phys. Chem. B 109, 21140–21144 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Persson F., Tegenfeldt J. O., DNA in nanochannels–directly visualizing genomic information. Chem. Soc. Rev. 39, 985–999 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Levy S. L., Craighead H. G., DNA manipulation, sorting, and mapping in nanofluidic systems. Chem. Soc. Rev. 39, 1133–1152 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Reymer A., Frykholm K., Morimatsu K., Takahashi M., Nordén B., Structure of human Rad51 protein filament from molecular modeling and site-specific linear dichroism spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 13248–13253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorfman K. D., Fulconis R., Dutreix M., Viovy J. L., Model of RecA-mediated homologous recognition. Phys. Rev. Lett. 93, 268102 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Wilhelmsson L. M., Westerlund F., Lincoln P., Nordén B., DNA-binding of semirigid binuclear ruthenium complex delta,delta-[μ-(11,11′-bidppz)(phen)(4)ru(2)](4+): Extremely slow intercalation kinetics. J. Am. Chem. Soc. 124, 12092–12093 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Feng B., Frykholm K., Nordén B., Westerlund F., DNA strand exchange catalyzed by molecular crowding in PEG solutions. Chem. Commun. (Camb.) 46, 8231–8233 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Almaqwashi A. A., et al. , Strong DNA deformation required for extremely slow DNA threading intercalation by a binuclear ruthenium complex. Nucleic Acids Res. 42, 11634–11641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berti D., Franchi L., Baglioni P., Luisi P. L., Molecular recognition in monolayers. Complementary base pairing in dioleoylphosphatidyl derivatives of adenosine, uridine, and cytidine. Langmuir 13, 3438–3444 (1997). [Google Scholar]
- 40.Matsuoka Y., Nordén B., Linear dichroism studies of nucleic acid bases in stretched poly(vinyl alcohol) film. Molecular orientation and electronic transition moment directions. J. Phys. Chem. 86, 1378–1386 (1982). [Google Scholar]
- 41.Matsuoka Y., Nordén B., Linear dichroism studies of nucleic acids. II. Calculation of reduced dichroism curves of A- and B-form DNA. Biopolymers 21, 2433–2452 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Matsuoka Y., Nordén B., Linear dichroism studies of nucleic acids. III. Reduced dichroism curves of DNA in ethanol-water and in poly(vinyl alcohol) films. Biopolymers 22, 1731–1746 (1983). [DOI] [PubMed] [Google Scholar]
- 43.Glaubiger D., Lloyd D. A., Tinoco I. Jr, Temperature-dependent optical properties of a torsional oscillator model for dinucleoside phosphates. Biopolymers 6, 409–414 (1968). [DOI] [PubMed] [Google Scholar]
- 44.Fornasiero D., “Studies of vibronic exciton interactions between pairs of chromophores,” PhD thesis, University of Adelaide, Australia (1981).
- 45.Schellman J. A., Flexibility of DNA. Biopolymers 13, 217–226 (1974). [DOI] [PubMed] [Google Scholar]
- 46.Wilson R. W., Schellman J. A., The flow linear dichroism of DNA: Comparison with the bead-spring theory. Biopolymers 17, 1235–1248 (1978). [DOI] [PubMed] [Google Scholar]
- 47.Parsegian V. A., Rand R. P., Rau D. C., Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc. Natl. Acad. Sci. U.S.A. 97, 3987–3992 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asakura S., Oosawa F., Interaction between particles suspended in solutions of macromolecules. J. Polym. Sci. 33, 183–192 (1958). [Google Scholar]
- 49.Zhang C., Shao P. G., van Kan J. A., van der Maarel J. R. C., Macromolecular crowding induced elongation and compaction of single DNA molecules confined in a nanochannel. Proc. Natl. Acad. Sci. U.S.A. 106, 16651–16656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano S., Karimata H., Ohmichi T., Kawakami J., Sugimoto N., The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 126, 14330–14331 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Miyoshi D., Nakamura K., Tateishi-Karimata H., Ohmichi T., Sugimoto N., Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 131, 3522–3531 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Spink C. H., Chaires J. B., Selective stabilization of triplex DNA by Poly(ethylene glycols). J. Am. Chem. Soc. 117, 12887–12888 (1995). [Google Scholar]
- 53.Moriyama R., Iwasaki Y., Miyoshi D., Stabilization of DNA structures with Poly(ethylene sodium phosphate). J. Phys. Chem. B 119, 11969–11977 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Stellwagen N. C., Stellwagen E., DNA thermal stability depends on solvent viscosity. J. Phys. Chem. B 123, 3649–3657 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Arnott S., Crystallography of DNA: Difference synthesis supports Watson-Crick base pairing. Science 167, 1694–1700 (1970). [DOI] [PubMed] [Google Scholar]
- 56.Pastor N., The B- to A-DNA transition and the reorganization of solvent at the DNA surface. Biophys. J. 88, 3262–3275 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berman H. M., Schneider B., “Nucleic acid hydration” in Oxford Handbook of Nucleic Acid Structure, Neidle S., Ed. (Oxford University Press, New York, 1999), pp. 295–312. [Google Scholar]
- 58.Kay E., Double-stranded DNA in methanol-ethanol-buffer solvent system. Biochemistry 15, 5241–5246 (1976). [DOI] [PubMed] [Google Scholar]
- 59.Zehfus M. H., Johnson W. C. Jr, Properties of P-form DNA as revealed by circular dichroism. Biopolymers 20, 1589–1603 (1981). [DOI] [PubMed] [Google Scholar]
- 60.Nordén B., Seth S., Tjerneld F., Renaturation of DNA in ethanol–methanol solvent induced by complexation with methyl green. Biopolymers 17, 523–525 (1978). [Google Scholar]
- 61.Kypr J., Kejnovská I., Renčiuk D., Vorlícková M., Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 37, 1713–1725 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee C. H., Mizusawa H., Kakefuda T., Unwinding of double-stranded DNA helix by dehydration. Proc. Natl. Acad. Sci. U.S.A. 78, 2838–2842 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui S., Yu J., Kühner F., Schulten K., Gaub H. E., Double-stranded DNA dissociates into single strands when dragged into a poor solvent. J. Am. Chem. Soc. 129, 14710–14716 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Limbach H. J., Holm C., Single-chain properties of polyelectrolytes in poor solvent. J. Phys. Chem. B 107, 8041–8055 (2003). [Google Scholar]
- 65.Gu B., Zhang F. S., Wang Z. P., Zhou H. Y., Solvent-induced DNA conformational transition. Phys. Rev. Lett. 100, 088104 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Phelps C., Lee W., Jose D., von Hippel P. H., Marcus A. H., Single-molecule FRET and linear dichroism studies of DNA breathing and helicase binding at replication fork junctions. Proc. Natl. Acad. Sci. U.S.A. 110, 17320–17325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.