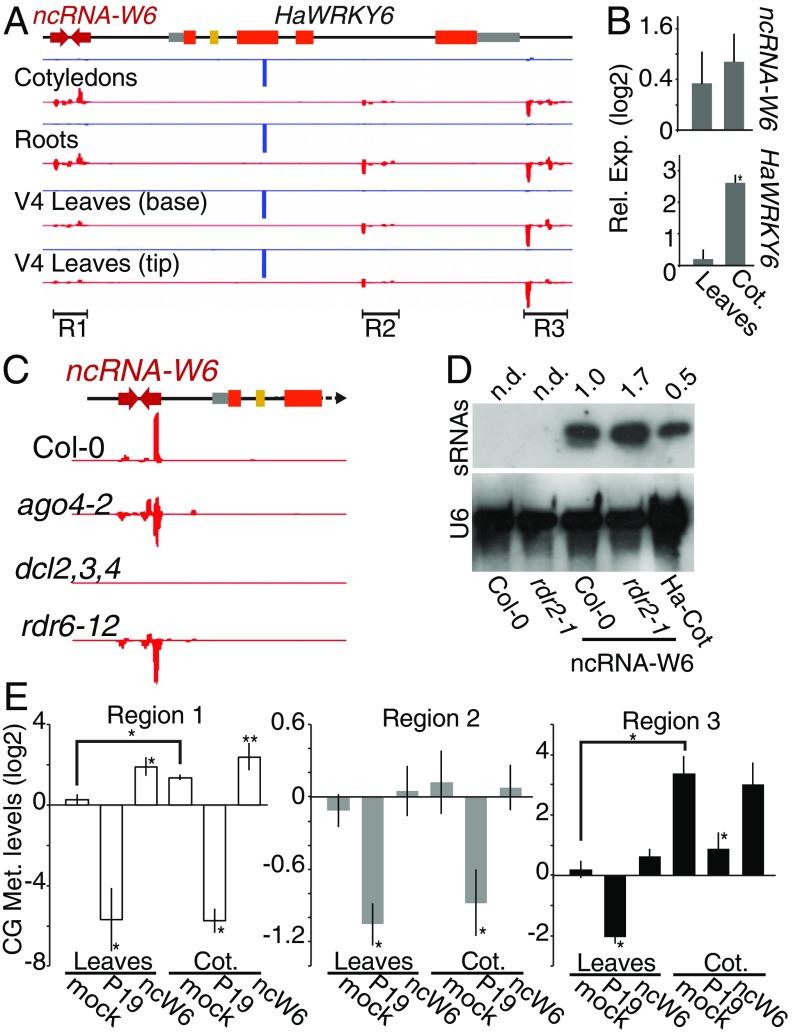
Fig. 2.
ncW6 generates epigenetically active sRNAs. (A) Alignment of sRNA sequencing reads to the HaWRKY6 locus in samples of different sunflower tissues; 24 nt (red) mapped to regions 1–3 and 21 nt (blue) corresponding to ha-miR396 reads mapped to the miRNA-target site in the gene. (B) ncW6 and HaWRKY6 transcript levels measured by RT-qPCR in sunflower Cots relative to leaves. (C) Alignment of sRNAs to a 35S::ncW6 construct transformed into Col-0, ago 4–2, dcl2,3,4, and rdr6-12 A. thaliana mutant plants. (D) RNA blots detecting ncW6-derived sRNAs in A. thaliana wild-type (Col-0) and rdr2-1 mutant plants control or transformed with 35S::ncW6 or sunflower Cots (Ha-Cot) as the positive control. U6 was used as a loading control and signal intensity calculated using ImageJ. Not detected signal (n.d.). (E) HpaII HaWRKY6 Chop-qPCR analysis of mock sunflower samples, plants overexpressing the silencing suppressor P19 (P19) as a sRNA decoy, or the ncW6. Digestion efficiency was quantified by qPCR with primers spanning restriction sites in the sRNAs mapping regions and normalized to an undigested region. Error bars show 2 × SEM, P values of less than 0.05 (**) or 0,01 (*) in a 2-tailed unpaired t test were considered significant.