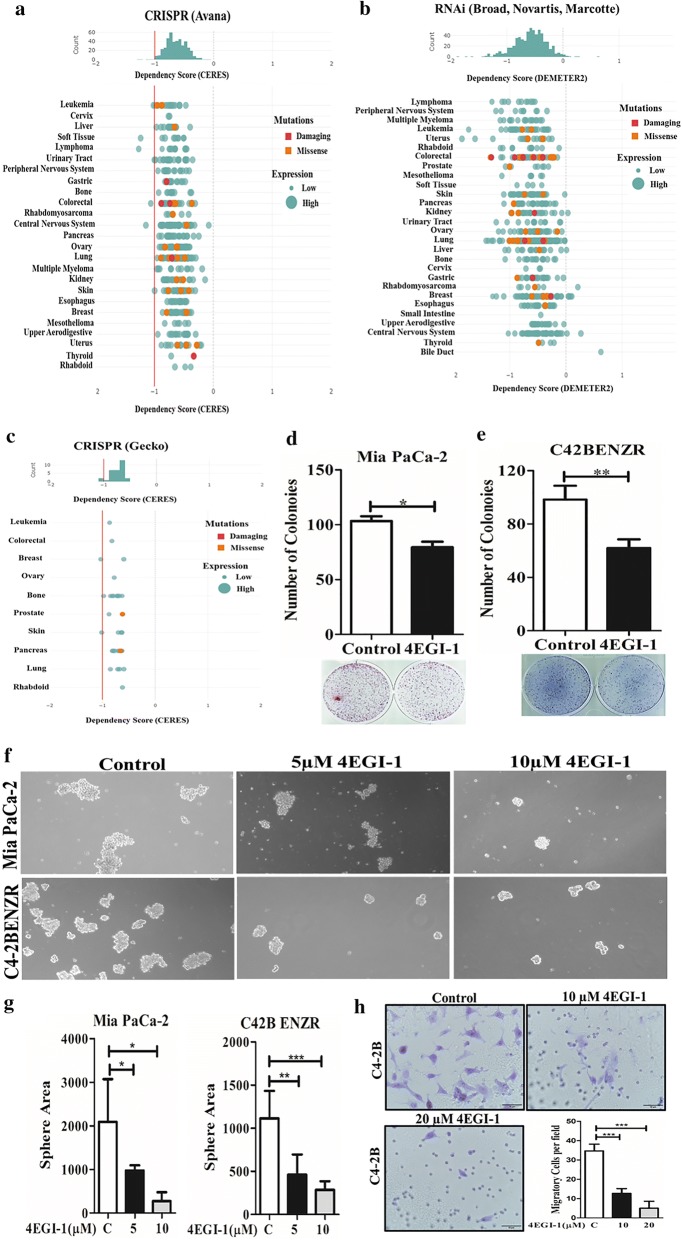
Fig. 6.
Critical requirement of EIF4G1 for cancer cell survival: Cancer cell survival analysis was done through DepMap portal. a CERES dependency score is based on data from a depletion assay for EIF4G1 by CRISPR-Cas9 (Avana) across cancers. b DEMETER2 score is based on shRNA (or siRNA) library containing multiple reagents designed to target the EIF4G1 gene in combined RNAi from Broad, Novartis, and Marcortte datasets. c CERES dependency score is based on data from a depletion assay for EIF4G1 by CRISPR-Cas9 (Gecko) across cancers. d, e Colony formation for Mia PaCa-2 (d) and C4-2BENZR (e) cells with control and treatment with EIF4G-EIF4E complex inhibitor (4EGI-1, 10 µM). The Colonies were counted using an NIH ImageJ software. Error bar represents ± SD. f Treatment with 4EGI-1 inhibitor (5/10 µM) significantly impaired the tumorosphere formation in for Mia PaCa-2 and C4-2BENZR cell lines. g The tumorosphere area was reduced significantly by treatment with 4EGI-1 inhibitor (5/10 µM). Error bar represents ± SD. h Representative Image (×40) and graph of Trans-well cell invasion assays on C4-2B cells. Treatment with 4EGI-1 inhibitor (10/20 µM) for 48 h, significantly inhibited cell invasion. Error bar represents ± SD