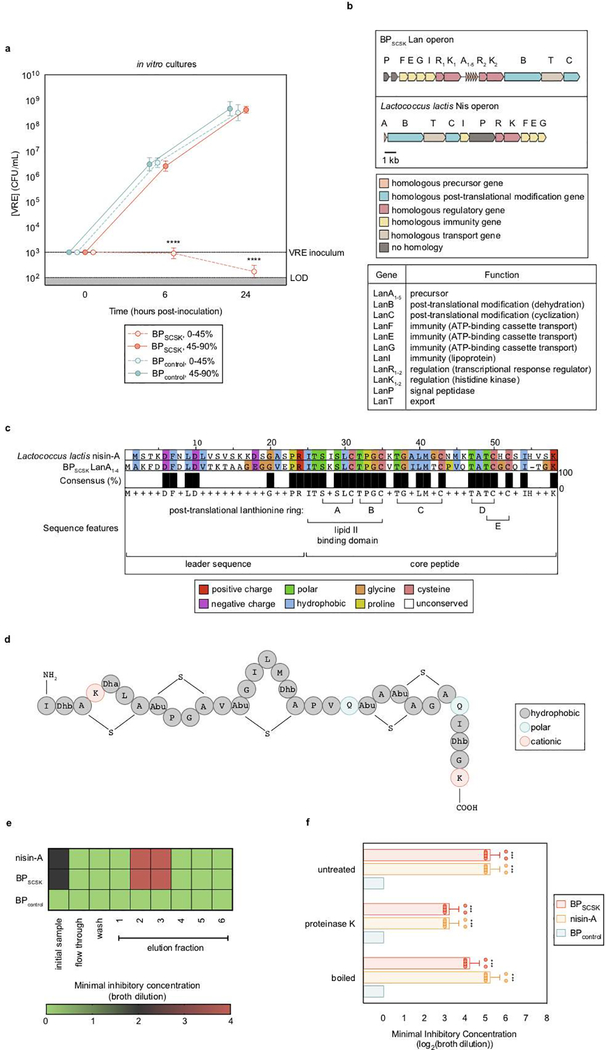
Extended Data Figure 5 |. BPSCSK encodes for a lantibiotic.
a, VRE was inoculated in media conditioned with BPSCSK or BPcontrol culture protein precipitate fractions (n = 8 biologically independent samples/2 independent experiments), and monitored for growth. b,c, BPSCSK was whole genome sequenced, assembled, and annotated. b, schematic comparing the lantibiotic operon discovered in BPSCSK’s genome to the nisin-A operon from Lactococcus lactis. Gene functions are based on the characterization of homologous genes in the nisin operon. c, amino acid sequence alignment comparing BPSCSK’s lantibiotic precursor (LanA1–4) and nisin-A precursor (NisA). Sequence features are based on the characterization of nisin. d, the molecular formula for the mature, post-translationally modified BPSCSK LanA1–4 lantibiotic with a predicted mass of 3152.45 Da. Dhb, dehydrobutyrine; Dha, dehydroalanine; Abu, alpha-aminobutyric acid. e, media conditioned with BPSCSK or BPcontrol culture protein precipitates, or commercial nisin-A, were incubated with proteinase K for 3 h at 37 °C, boiled at 100 °C, or left untreated. The treated protein precipitate (n = 8 biologically independent samples/4 independent experiments) was serially diluted and VRE was inoculated and cultured for 24 h. The minimal inhibitory concentration (MIC) was the highest mean dilution where VRE inhibition was observed. c, Proteins were precipitated from BPSCSK or BPcontrol, or nisin-A spiked cultures and applied to a SP sepharose column. Each fraction was serially diluted and VRE was inoculated and cultured for 24 h to determine the MIC (n = 4 biologically independent samples/4 independent experiments). VRE (ATCC 700221) was used in experiments (a,e,f). All statistical analyses were performed using the Mann-Whitney rank sum test (two-tailed) comparing experimental conditions to a negative control. *** p-value < 0.001, ****p-value < 0.0001. Data points (geometric mean), error bars (geometric s.d.) (a); mean (e); center values (geometric mean), error bars (geometric s.d.) (f).