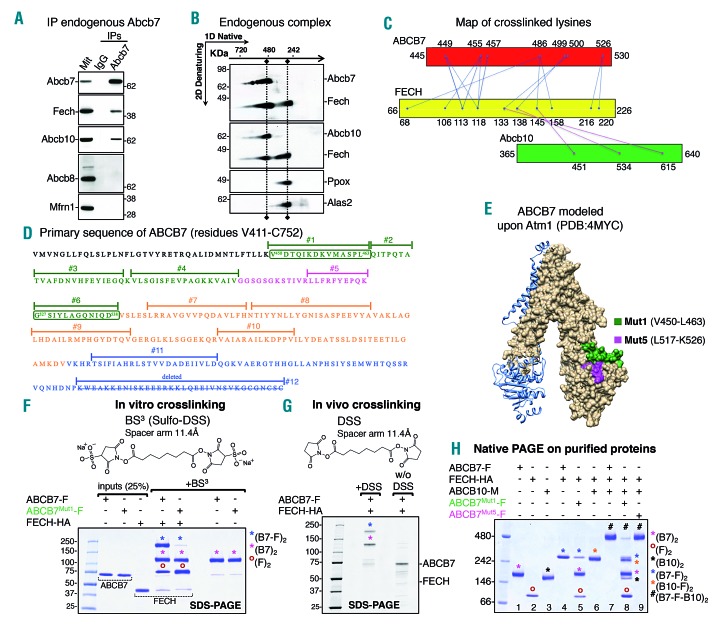
Figure 5.
ABCB7 forms a complex with ferrochelatase and Abcb10 through direct interaction revealed by crosslinking. (A) Immunoprecipitation (IP) of endogenous Abcb7 in G1E-ER4 cells 30 h after induction of differentiation showed formation of a complex between Abcb7, Fech and Abcb10 that did not include Abcb8 or Mfrn1. (B) Native/two-dimensional (2D) sodium dodecylsulfate polyacrylamide gel electropheresis (SDS-PAGE) analysis on mitochondrial lysates from G1E-ER4 cells showed co-migration of Abcb7, Fech and Abcb10 in a single complex of an approximate molecular weight of 480 kDa. A second distinctive pool of Fech co-migrated with the heme-synthesizing enzymes Ppox and Alas2, indicating formation of a complex at 250 kDa. (C) Map showing the crosslinked sites in the ABCB7/FECH/Abcb10 complex. Inter-subunit crosslinks between ABCB7 and FECH are in blue, and inter-subunit crosslinks between FECH and Abcb10 are in magenta. The lysine residues crosslinked by DDS in the protein sequences are represented by dots. (D) Primary sequence of the C-terminal domain of ABCB7 between residues Val 411 and Cys752. Peptide sequences that were substituted by alanines to test their involvement in the interaction with Fech are highlighted in different colors. (E) Three-dimensional structure of ABCB7 modeled on the structure of S. cerevisiae Atm1 (PDB:4MYC3) using Swiss-Model.49 The last 44 amino acid residues of ABCB7 are missing from the structure because yeast lacks these terminal residues. One of the two protomers of ABCB7 in the dimeric structure is represented in the surface-mode and the green and magenta sequences in the C terminus of ABCB7 indicate the peptide sequences subjected to alanine scanning mutagenesis in Mut1 and Mut5, respectively, to assess their involvement in the interaction with FECH. (F) Coomassie staining of inputs and in vitro crosslinked products on SDS-PAGE. Magenta asterisks denote dimers of ABCB7 wildtype (WT) or the mutant (Mut1) in which amino acid residues between Val450 and Leu463, involved in binding FECH, were replaced by alanines. Blue asterisks indicate the tetrameric ABCB7-FECH complex. Brown circles indicate FECH dimers. (G) SDS-PAGE analysis after in vivo crosslinking on mitochondria isolated from cells co-transfected with ABCB7-F and FECH-HA, followed by anti-FLAG immunoprecipitation. (H) Native PAGE on purified ABCB7-FLAG, FECH-HA and ABCB10-Myc proteins shows that both ABC transporters dimerized when loaded individually (lanes 1 and 3; magenta and black asterisks correspond to dimers of ABCB7 and ABCB10, respectively). Both ABCB7 and ABCB10 were able to interact physically with dimers of FECH (lanes 4 and 6; blue and orange asterisks denote the hetero-tetrameric ABC transporter-FECH complexes). ABCB7, FECH and ABCB10, when combined together in vitro, formed a multiprotein complex with a 2:2:2 stoichiometry consisting of dimers of each of the components (in lane 7, the complex denoted with the # symbol). ABCB7Mut1, in which amino acid residues between Val450 and Leu463 were replaced by alanines, was unable to interact with FECH (lane 5) and formation of the hexameric complex was disrupted (lane 8), whereas Mut5 showed no defect in interacting with FECH (lane 9). Brown circles indicate FECH dimers (lane 2). (A, B, n=5; F, G and H, n=3).