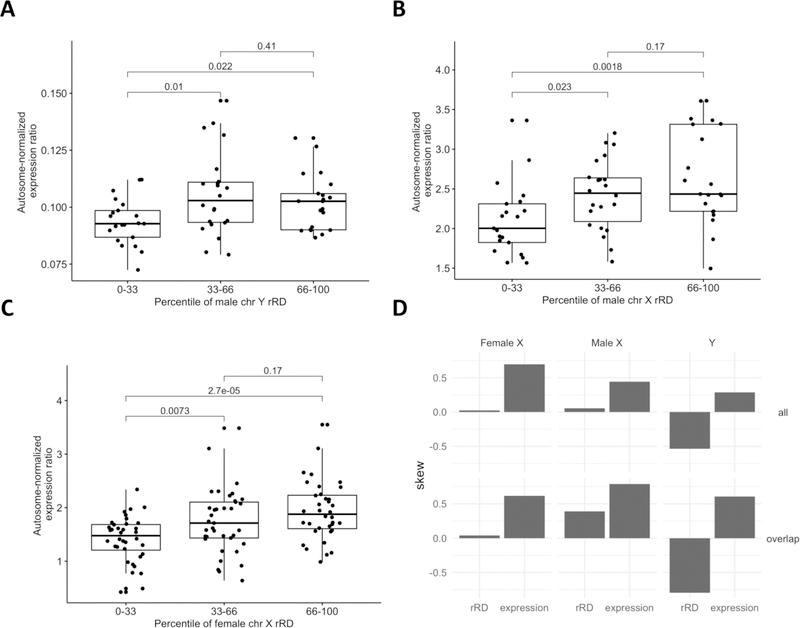
Figure 7: Relationship between sex chromosome rRD and proportion of gene expression attributable to that chromosome.
A, B, C) Here, rRD was used instead of z-score because rRD exhibits improved overall correlation with mLRR. Further, a single DNA extraction kit (QIAamp) was used to extract both the WGS and RNAseq data, therefore negating the need for kit-specific correction. Briefly, a measure of the proportion of gene expression attributable to the sex chromosomes was defined as the ratio of the median of the top 20 highly expressed genes on the sex chromosome in question (or top 13 on the Y) to the median of the top 440 highly expressed genes across all autosomes (20 from each autosome, see Methods, section 2.7.1). An increase in total amount of a sex chromosome in the sample would be expected to lead to an increase in the proportion of that chromosome’s gene expression relative to the autosomes. After correcting for RIN score, PMI and RNA sequencing batch, correlation between rRD and Y expression ratio and X expression ratio was observed in the male Y and female X chromosomes (Y chr: r = 0.29, p = 2.42e-02, female X chr: r = 0.36, p =1.2e-04; male X chr: r = 0.23, p = 6.5e-02). P-values from ANOVA are shown on graphs for ease of visualization. D) Distribution in skew across all WGS and RNAseq data. “all” signifies the skew in all available data for each data type, not just those individuals common to WGS and RNA-seq analyses. “Common” signifies the skew in the data common to both WGS and RNA-seq analyses. In the female and male X chromosomes, positive skew is observed at both the RNA and DNA level. In contrast, in the Y chromosome, conflicting skew is observed at the DNA and RNA level.