Figure 1.
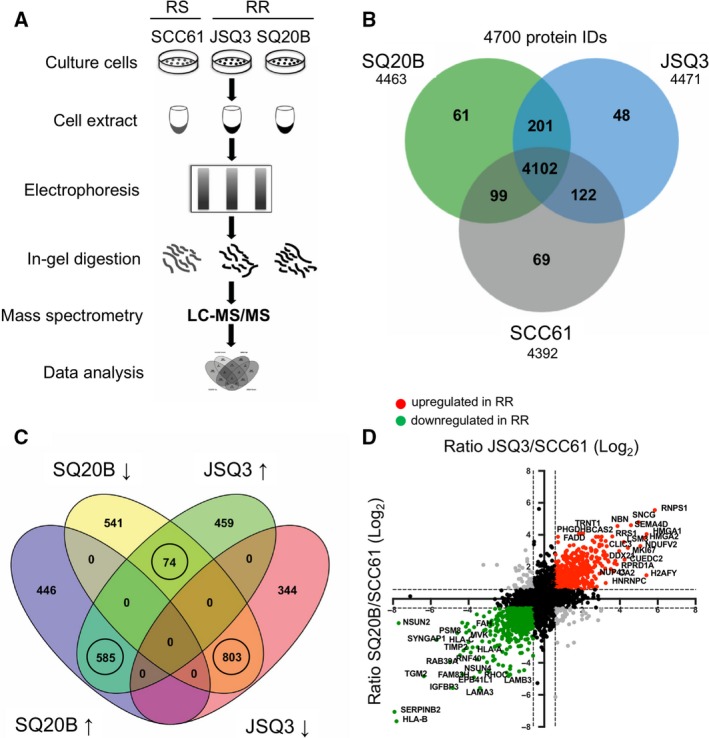
Proteomic analysis reveals a distinct radioresistant HNSCC cell proteome. (A) Protocol schematic for mass spectrometry assay. Whole‐cell protein lysates were prepared from SCC61 (radiosensitive), JSQ3 (radioresistant), and SQ20B (radioresistant) HNSCC cell lines. Proteins were separated by gel electrophoresis, digested with trypsin, and analyzed via LC‐MS/MS. (B) Venn diagram showing proteins uniquely identified in one or two cell lines, or proteins shared between all cell lines. 201 proteins were shared by the two radioresistant cell lines (SQ20B and JSQ3) and 69 proteins were unique to radiosensitive cells (SCC61), while 4102 proteins were shared by all three cell lines. (C) Venn diagram showing distribution of up‐ and downregulated proteins shared between the two radioresistant cell lines, SQ20B and JSQ3, compared to the radiosensitive cell line, SCC61. In general, most proteins found in both radioresistant cell lines were similarly regulated, with 585 shared proteins upregulated and 803 shared proteins downregulated. Only 74 proteins displayed variable regulation. Only circled groups were considered for further analysis. (D) Representation of quantitative mass spectrophotometric intensity ratios of SQ20B/SCC61 (x‐axis) and JSQ3/SCC61 (y‐axis) shown on log2 scale. A 1.5‐fold (0.58 log2) change was considered statistically significant. Upregulated proteins, red. Downregulated proteins, green. Variably regulated proteins, gray; nonsignificant proteins, black. The data show a linear trend, with proteins up‐ or down‐regulated in the SQ20B vs. SCC61 cell line tending to be similarly regulated in JSQ3 vs. SCC61 samples.
