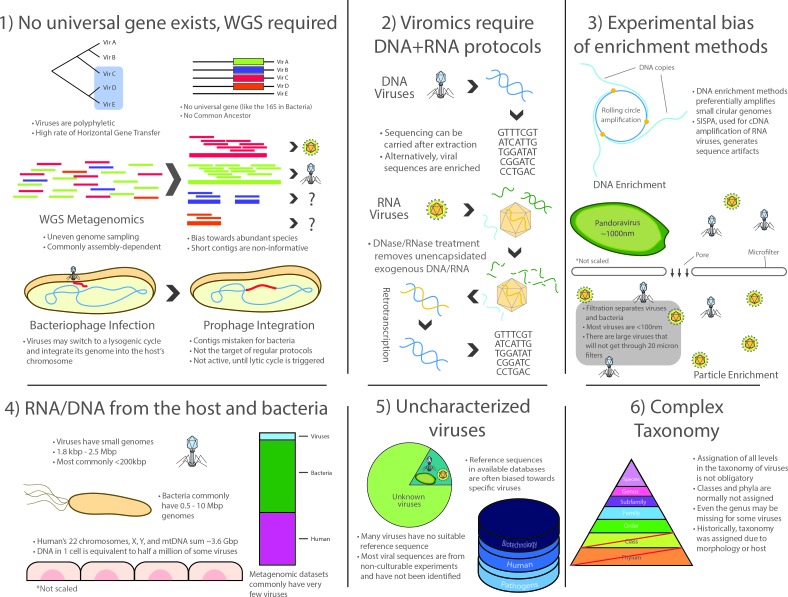
Figure 1. FIGURE 1: Challenges in the study of the virome.
The study of viral communities faces numerous experimental limitations that are inherent to the study of viral particles. As viruses are polyphyletic, viromic-level phylogenies are often unreliable, further complicated by the lack of universal gene makers, a high rate of horizontal gene transfer, and the lack of a common ancestor. Viral metagenomics thus rely on the WGS framework which is commonly dependent of (yet, not restricted to) sequence assembly. This derives in additional challenges such as the formation of chimeric contigs, underrepresented fragments and species, and other general assignation issues. Additional hurdles are presented in viral RNA workflows as unencapsidated nucleic acids must be removed prior to RNA extraction, following retrotranscription. Additional biases may be introduced by enrichment of viral particles (filtration is shown as an example) or by amplification techniques for optional DNA enrichment, such as the rolling circle amplification. Niche ecology may also affect the recovery of viral sequences, as nucleic acids from the host and its bacteria are abundant in sample preparation and are often sequenced unintentionally. Moreover, suitable viral reference sequences are often missing from public sequence databases. Plus, several of those that exist lack a proper functional characterization or taxonomic assignation and databases are commonly biased towards specific species with pathogenic or biotechnological potential. Furthermore, current viral taxonomy is convoluted, with a large proportion of viruses having inaccurate or missing taxonomic labels, often derived from morphology or feature-based assignations. Together, these challenges have hindered the advance of viromics in the past decade, representing a hefty entry barrier for the scientific community due to the increased time, expertise, and resources that are required by these studies with respect to their bacterial counterpart.