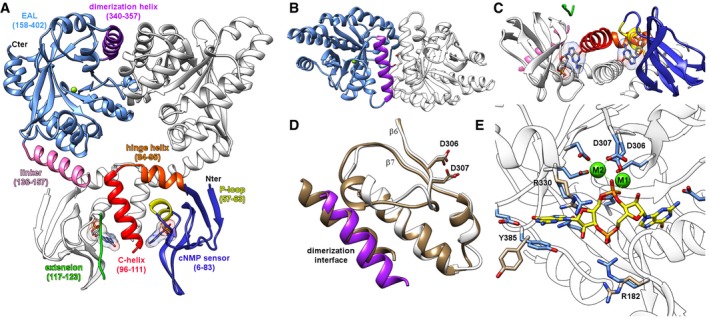
Dimer of full‐length cGMP‐bound Bd1971; one monomer colored individually by subdomain, the opposing monomer colored white. Ligand represented in stick form, EAL active site Mg2+ as green sphere.
View from top of that in (A), detailing EAL region of dimer interface.
View from opposing side, detailing C‐helix dimer interface at cNMP domain.
The Bd1971 EAL dimerization interface (purple, white) matches that of a canonical, active dimer from the well‐characterized T. denitrificans EAL (tan, PDB 2r6o); the catalytic Asp pairs of the two enzymes adopt a similar conformation (stick form, D306/307 from Bd1971, D646/647 from 2r6o).
Structure of the cyclic‐di‐GMP bound complex of Bd1971 (ligand C represented in yellow, bound Ca2+ ions as green spheres). Residues R182, R330, and Y385 display the largest shift between the apoenzyme (tan) and bound (blue) states, R182 complexing the cyclic‐di‐GMP phosphate, and Y385 stacking with a guanine base.