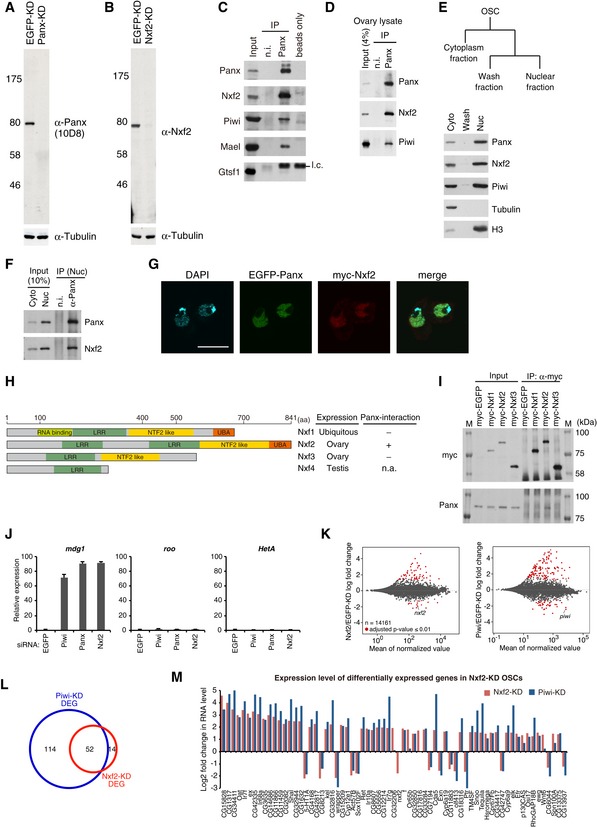
Western blotting (WB) shows the specificity of anti‐Panx monoclonal antibodies raised in this study. Tubulin was used as a loading control.
WB shows the specificity of anti‐Nxf2 monoclonal antibodies raised in this study.
Immunoprecipitation (IP) from OSC lysate using anti‐Panx antibody, followed by WB of Panx, Nxf2, Piwi, Mael, and Gtsf1. IP‐WB was performed under the same conditions as the silver staining described in Fig
1A. Mouse immunoglobulin G (IgG) (n.i.) was used for control IP. l.c.: light chain from the antibody.
IP from fly ovary lysate using anti‐Panx antibody, followed by WB of Panx, Nxf2, and Piwi.
Scheme of preparation of cytoplasmic and nuclear fractions of OSCs. WB using Panx, Nxf2, Piwi, Tubulin, and histone H3 antibodies shows the level of each protein in the separate fractions. Tubulin was detected in the cytoplasmic fraction, where histone H3 was enriched in the nuclear fraction.
IP from nuclear OSC lysate using anti‐Panx antibody, followed by WB.
Immunofluorescence of OSCs co‐transfected with EGFP‐Panx‐ and myc‐Nxf2‐expressing vectors, using myc antibody (red). EGFP is detected as Panx signal, and DAPI staining (blue) shows the location of nuclei. EGFP‐Panx and myc‐Nxf2 co‐localize in the nucleus. Scale bar: 10 μm.
Schematic of NXF variants. LRR: leucine‐rich repeat, NTF2‐like: nuclear transport factor 2‐like domain, UBA: ubiquitin‐associated domain (left panel). Nxf1 is ubiquitously expressed, while Nxf2 and Nxf3 are almost exclusively expressed in ovary, and Nxf4 is specifically expressed in testis. The interaction between Nxf4 and Panx was not analyzed, since Nxf4 expression is limited to testis, indicated as n.a. (not available) (right panel).
IP from lysate of OSCs expressing myc‐tagged NXF variants, followed by WB using anti‐myc and anti‐Panx antibodies. M indicates protein markers. The results are summarized in the right panel in (H). Among NXF variants, only Nxf2 can interact with Panx.
RNA levels of mdg1, roo, and HetA were quantified by qRT–PCR upon depletion of EGFP (control), Piwi, Panx, or Nxf2. Expression levels are normalized by the expression of RP49. Error bars represent SD (n = 3). Piwi–piRNA‐targeted TE was specifically de‐silenced by Piwi‐, Panx‐, and Nxf2‐KD, confirming the results of RNA‐seq.
MA plot of RPKM values (log10 scale) for mRNAs in the indicated KD samples, based on RNA‐seq. Differentially expressed genes (DEGs) are in red.
Venn diagram displaying the number of DEGs upon depletion of Nxf2 (red) or Piwi (blue).
Log2 fold changes in mRNA levels of 67 DEGs in Nxf2‐KD OSCs. mRNA levels calculated from RNA‐seq data are used for Nxf2‐ and Piwi‐KD OSCs. Most mRNAs whose expression was altered upon Nxf2‐KD were also affected by Piwi‐KD, suggesting that Nxf2 does not globally affect mRNA levels.