Abstract
Hypoxia is a tumorigenesis-related microenvironment change which usually occurs in the earliest stage of prostate cancer (PCa) development. Accumulating evidence has demonstrated that hypoxia/hypoxia-inducing factor (HIF) is involved in the induction of epithelial-mesenchymal transition (EMT) and increased metastatic potential in PCa. However, the mechanism by which hypoxia/HIF regulates EMT remains unclear. In the present study, we demonstrated the molecular mechanisms of hypoxia-induced EMT in PCa, focusing on HIF-1α/Forkhead box M1 (FoxM1) signaling pathway. PCa PC3 and DU145 cell lines were used as the model system in vitro. Our data revealed that hypoxia induced EMT in PCa cells. Bioinformatics analysis identified the possible association between HIF-1α and FoxM1. Additionally, FoxM1 was significantly associated with PCa development and Gleason scores of PCa. Exposure to hypoxia resulted in the increased expression of HIF-1α and FoxM1. Genetic knockdown FoxM1 abolished hypoxia-induced EMT in PCa, while exogenous overexpression of FoxM1 facilitated hypoxia-induced EMT. Furthermore, the increase of FoxM1 during hypoxia was due to the transcriptional regulation on the FoxM1 promoter by HIF-1α. We also confirmed the binding site of HIF-1α on the FoxM1 promoter by different lengths promoter sequences. These findings provide new insights into how EMT is regulated in PCa under hypoxic stress. It is worthwhile to investigate in future that inhibition of FoxM1 as a potential target may be an effective therapeutic strategy against PCa.
Keywords: hypoxia-inducing factor-1, Forkhead box M1, hypoxia, epithelial-mesenchymal transition, prostate cancer
Introduction
Prostate cancer (PCa) is the most common cancer in men aged ≥65 years with an average age of ~66 at the time of diagnosis (1). PCa ranks the third leading cause of male cancer-associated mortality with an estimated 31,620 deaths in 2019 in the USA. Metastasis is the main cause of PCa-related mortality, in which cancer cells spread to bladder, bone, retroperitoneal lymph nodes, spinal cord and other areas (2,3). About 40% of localized PCa patients relapsed after initial therapy (4) and they usually succumb due to cancer metastasis or drug resistance. Thus, it is a great challenge to determine the underlying mechanism of PCa progression, recurrence and metastasis to improve the outcomes of treating PCa.
Hypoxia is a tumorigenesis-related microenvironment change which usually occurs in the earliest stage of PCa development (5). In response to decreasing oxygen availability, the activity of hypoxia-inducible factors (HIFs) in cells increase and also mediate various transcriptional changes (6). HIF heterodimers comprise HIFα and HIFβ, the former is sensitive to oxygen and the latter is a constitutively expressed subunit. Under hypoxic conditions, HIF-1α dimerizes with HIF-1β and translocates into nucleus to bind to the hypoxia-responsive element, which is the specific sequence present in the promoter of several hypoxia-dependent target genes (7). Mechanistically, hypoxia was demonstrated to affect the invasive and migratory behavior of PCa cells via epithelial-to-mesenchymal transition (EMT), a trans-differentiation of cells for the acquirement of plasticity and increased mobility, a process which alters the metastatic potential of cancer cells (8). It is widely reported that hypoxia is the inducer of the EMT process in various epithelial cancers, such as PCa, ovarian carcinoma, lung cancer and hepatocellular carcinoma (9–11), thus facilitating tumor cell survival and resistance to chemo or radio-therapies (12,13). Hypoxia-induced EMT is characterized by a decrease in epithelial gene expression, such as E-cadherin and β-catenin and an increase in mesenchymal associated gene expression, such as N-cadherin and vimentin (14). Hypoxia can also activate downstream transcription factors such as Smads, SNAIL, SLUG, and TWIST, and inhibits the expression of E-cadherin (15). Recently, it was reported that chronic hypoxia-induced SLUG promotes the EMT of PCa cells by activating expression of Ephrin-B1 (16); however, the detailed mechanisms leading to the induction of EMT by hypoxia remains largely unknown.
Forkhead box M1 (FoxM1) is a classic proliferation- associated transcription factor belonging to the family of Forkhead box (Fox) protein, which consists of a conserved forkhead DNA-binding domain, an N-terminal repressor domain, and a C-terminal transactivation domain (17). In addition to cell proliferation, FoxM1 is also involved in cell cycle regulation, angiogenesis, invasion, metastasis and EMT (18,19). FoxM1 has been reported to be upregulated and of prognostic significance in several malignancies including breast cancer, colorectal cancer, lung cancer, melanoma and PCa (20–25). Over the past few decades, understanding of the function and regulation of FoxM1 has notably improved, providing novel insights into the roles of this transcription factor in cancer development and progression. Additionally, certain small molecule inhibitors that target FoxM1 have promising potential as therapeutic drugs against PCa and have been receiving great attention by urologists and patients. There is thus increasing interest in elucidating the regulatory mechanisms of FoxM1 in PCa.
In our study, we reported that FoxM1 was transcriptionally regulated by HIF-1α in PCa. FoxM1 was upregulated in PC3 and DU145 PCa cells under hypoxic conditions, which were associated with EMT induction and enhanced invasive ability. This process could be inhibited by the suppression of HIF-1α activity. Furthermore, HIF-1α binding sites were illustrated and mapped to the promoter of FoxM1. Thus, we concluded that EMT of hypoxic PCa cells is mediated by the transcriptional regulation of FoxM1 by HIF-1α.
Materials and methods
Reagents and antibodies
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), penicillin and streptomycin cocktail were purchased from HyClone (GE Healthcare). Primary antibodies against β-actin (cat. no. 3700), E-cadherin (cat. no. 3195), vimentin (cat. no. 3932), FoxM1 (cat. no. 5436), and HIF-1α (cat. no. 3716) were purchased from Cell Signaling Technology; the dilutions for primary antibody es were 1:1,000. YC-1, proteinase and phosphatase inhibitors cocktail were from Sigma-Aldrich (Merck KGaA). Transwell mini-cells were obtained from EMD Millipore and Matrigel was purchased from BD Medical Technology (BD Biosciences). Nitrocellulose membranes and Enhanced Chemiluminescence reagents were purchased from Bio-Rad Laboratories, Inc.
Cell culture
PC3 and DU145 cell lines were purchased from the American Type Culture Collection. Both cell lines were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were incubated in an atmosphere of 95% humidity at 37°C, 5% CO2. Culture medium was replaced with fresh medium every other day or according to experimental designs. For hypoxia, cells were plated and adhered overnight, then cells were grown in a hypoxia incubator at 37°C in 1% O2, 5% CO2 and 94% N2 for different time periods (24, 48 and 72 h). For YC-1 treatment, cells were pretreated with 50 µM YC-1 for 24 h at 37°C, 5% CO2 and then exposed to normoxic or hypoxic conditions. To detect the changes of HIF-1α and FoxM1 in both mRNA and protein levels under hypoxic condition, cells were plated and adhered overnight under normoxic condition and then exposed to hypoxic condition for different time periods (0, 6, 12 and 24 h).
Lentivirus preparation, siRNA, and transfection
PLKO.1 lentiviral vector was used to package encoding short hairpin RNAs (shRNAs) of FoxM1 with the sequence, 5′-GGAAATGCTTGTGATTCAACA-3′. To generate the lentivirus, 8 µg PAX2 packaging vector, 2 µg VSV-G and 8 µg the aforementioned plasmids or empty vectors as the negative control were co-transfected into 90% confluent 293T cells (purchased from the American Type Culture Collection) in 10 cm plates. Lentivirus expressing FoxM1 was produced using pcDNA3-FoxM1 plasmid, which was sub-cloned into LV5 vectors according to the manufacturer's protocols (Shanghai GenePharma Co., Ltd.). The pcDNA3-FoxM1 vector was generated by the human FoxM1 coding region cDNA subcloning into the pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) plasmid. The empty vectors were used as the negative control. The transfection reagent used for the above protocols was Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. Cells were infected by the virus with multiplicity of infection of 40 then selected and maintained in 2–3 µg/ml puromycin for subsequent experiments. Small interfering RNA targeting HIF-1α was designed and synthesized by Shanghai GenePharma Co., Ltd. The sequence of HIF-1α siRNA was as follows: siHIF1α: 5′-AGCACUACUUCGAAGUGGCTT-3′. Cells were transfected with X-tremeGene siRNA transfection reagents (Roche Diagnostics GmbH) for 2–3 days, and harvested for subsequent experiments.
Matrigel migration and invasion assay
Cells were seeded onto 6-well plates and cultured to 60% confluence. Then, cells were treated with or without hypoxia for 24 h. Cells were digested with Trypsin-EDTA Solution (0.25% trypsin, 0.02% EDTA) and centrifuged at 160 × g, room temperature for 5 min. 50,000 cells were seeded onto a Transwell chamber (Corning, Inc.) without Matrigel (migration assay) and 8×104 cells were seeded onto a Transwell chamber with Matrigel (invasion assay) in 200 µl serum-free medium. For invasion assay, Matrigel was diluted with serum-free medium (1:4) and coated by 40 µl onto the Transwell chamber then put in 37°C for 1 h; 1 ml medium containing 10% FBS was added to the lower chamber (24-well plate) as a chemoattractant. After incubation at 37°C for 20 h, non-migrated or non-invaded cells on the upper surface of the filter were removed with a cotton swab. Cells invaded to the underside of the filter membrane were stained by 0.1% crystal violet at room temperature for 10 min and the cell numbers were calculated statistically in five random fields using an inverted light microscope (magnification, ×200).
Western blot analysis
Cells were prepared with radioimmunoprecipitation assay buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 1% NP-40, and 0.5% sodium deoxycholate). Protein was separated by 12% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked in 5% skim milk at room temperature for 1 h and then incubated with primary antibodies at 4°C overnight. Following washing with TBST, membranes were incubated with Goat anti-rabbit (cat. no. 926-32211) or goat anti-mouse secondary antibodies (cat. no. 926-68070) (LI-COR Biosciences) diluted at 1:3,000 in 5% skimmed milk at room temperature in the dark for 1 h. Following washing with TBST, membranes were visualized by an Odyssey infrared imaging system (Li-Cor Biosciences). The relative intensity of each band was determined by using Glyko BandScan software version 4.0 (Glyko Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Cells were harvested with TRIzol reagent (Thermo Fisher Scientific, Inc.) to extract total RNA. Then, the lysates ware reverse-transcribed into cDNA using a PrimeScript™ RT reagent kit (Takara Bio, Inc.) according to the manufacturer's protocols. cDNA was employed for qPCR using a CFX96 real-time PCR system (Bio-Rad Laboratories, Inc.) with SYBR®-Green PCR Master Mix (Takara, Dalian, China) to determine the expression of target genes. The thermocycling conditions were defined as follows: 95°C 10 min, 1 cycle; 95°C 10 sec, 60°C 30 sec, 72°C 30 sec, 40 cycles; 72°C 10 min, 1 cycle. Relative gene expression was calculated by the 2−ΔΔCq method (26). The primers used were: β-actin, forward, 5′-AAGGATTCCTATGTCGGC-3′ and reverse, 5′-CTTCATGATGGAGTTGAAGGT-3′; HIF-1α, 5′-AGCTTGCTCATCAGTTGCCA-3′ and reverse, 5′-CCAGAAGTTTCCTCACACGC-3′; FoxM1, 5′-GGAGCAGCGACAGGTTAAGG-3′ and reverse, 5′-GTTGATGGCGAATTGTATCATGG-3′.
Dual-luciferase activity assay
FoxM1 promoter report plasmid pGL3-FoxM1 was generated by inserting different lengths promoter fragments of FoxM1 (−1,000/+40, −878/+40, −300/+40, −220/+40, −50+40) into the pGL3-basic plasmid (Shanghai GenePharma Co., Ltd.). The vector construct was validated by sequencing. PC3 and DU145 cells under normoxic or hypoxic conditions were cotransfected with ERE-TK-Luc, pGL3-basic, and pGL3-FoxM1 using X-tremeGENE HP DNA transfection reagent (Roche Diagnostics). Then, a dual-luciferase activity assay was carried out using the Dual Luciferase Assay kit (Promega Corporation) following the manufacturer's instructions. Each data point used three wells of cells and the results were calculated statistically.
Bioinformatics analysis
The generate a heatmap for analysis, the RNA-sequencing data of PCa in count format was downloaded from The Cancer Genome Atlas (TCGA: http://cancergenome.nih.gov/). Then, we sorted the data according to the level of HIF1A expression. The top 50 samples with high expression of HIF1A and the bottom 50 samples with low expression of HIF1A were extracted. Furthermore, differentially expressed genes (DEGs) were identified by edgeR package (v3.26.5, http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) (27,28) using R software; P<0.05 and fold change >2 were considered as statistically significant. The expression of 50 DEGs were presented in the heatmap.
In addition, the expression of FoxM1 in TCGA prostate normal and cancer samples with the Gleason score was presented as a boxplot, which was analyzed using the UALCAN online tool (http://ualcan.path.uab.edu/analysis.html) with P<0.01 among groups. Gene Ontology (GO) and pathway enrichment analyses were performed on the top 200 upregulated DEGs using Metascape (http://metascape.org/gp/) (29). Moreover, the RNA-sequencing data of PCa in FPKM format was downloaded from TCGA, after log2(x+1) normalization, the HIF1A and FOXM1 expression were extracted; Pearson's correlation was performed using R software.
Statistical analysis
GraphPad Prism 7 software (GraphPad Software, Inc.) was used for the statistical analysis. The difference between two groups was analyzed by a Student's t-test. Comparisons between multiple groups were performed by one-way analysis of variance, followed by Dunnett's post-hoc test in which one group was compared against all the other groups or a Tukey's multiple comparison test in which pairwise comparisons between all groups were performed. Data were presented as mean ± standard deviation of three replicates. P<0.05 was considered to indicate a statistically significant difference.
Results
Hypoxia results in the induction of EMT with upregulation of HIF-1α and FoxM1 in PCa cells
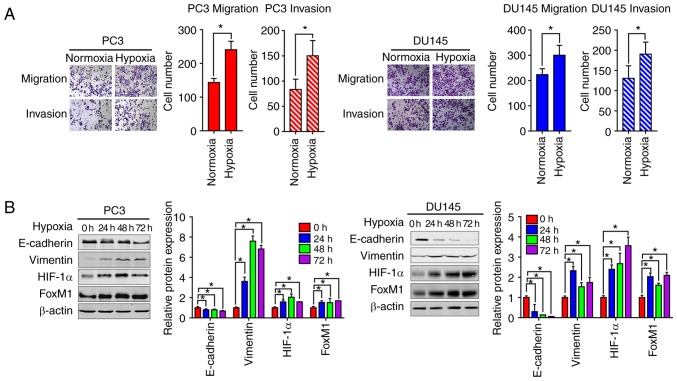
We firstly investigated the effects of hypoxia on the EMT of PCa cells. Transwell migration and invasion assays were performed to assess the migration and invasive abilities of PC3 and DU145 cells. Compared with the normoxic control groups, the number of migrated and invasive cells was significantly higher under hypoxic conditions compared with normoxic conditions. This indicated that hypoxia could increase the migration and invasion abilities of PC3 and DU145 cells (Fig. 1A). Furthermore, the expression of epithelial and mesenchymal phenotypic markers was determined by western blotting analysis. As presented in Fig. 1B, compared with the normoxic conditions, the expression of E-cadherin was significantly decreased, while that of vimentin was significantly increased in PC3 and DU145 cells under hypoxic conditions; the highest levels of vimentin were observed at 48 h. Increased expression of HIF-1α and FoxM1 protein levels were also found under hypoxic conditions.
Figure 1.
Hypoxia induces EMT in PC3 and DU145 prostate cancer cells with the upregulation of HIF-1α and FoxM1. (A) The migration and invasion were tested by Transwell invasion assay in PC3 (left panel) and DU145 (right panel) cells. Cells were placed onto the upper chamber without or with Matrigel under normoxic or hypoxic conditions (24 h) and the cell images were obtained (magnification, ×200). Quantification analysis is shown. (B) PC3 and DU145 cells were cultured under normoxic or hypoxic conditions for different time periods (0, 24, 48 and 72 h), then the protein expression levels of EMT markers, including E-cadherin, vimentin, HIF-1α and FoxM1 were detected by western blotting. Similar results were observed in two additional experiments. Data are presented as the mean ± standard deviation of three replicates. *P<0.05 vs. control. EMT, epithelial-mesenchymal transition; FoxM1, Forkhead box M1; HIF-1α, hypoxia-inducible factor 1α.
Bioinformatics analysis of potential correlation between HIF-1α and FoxM1 in PCa
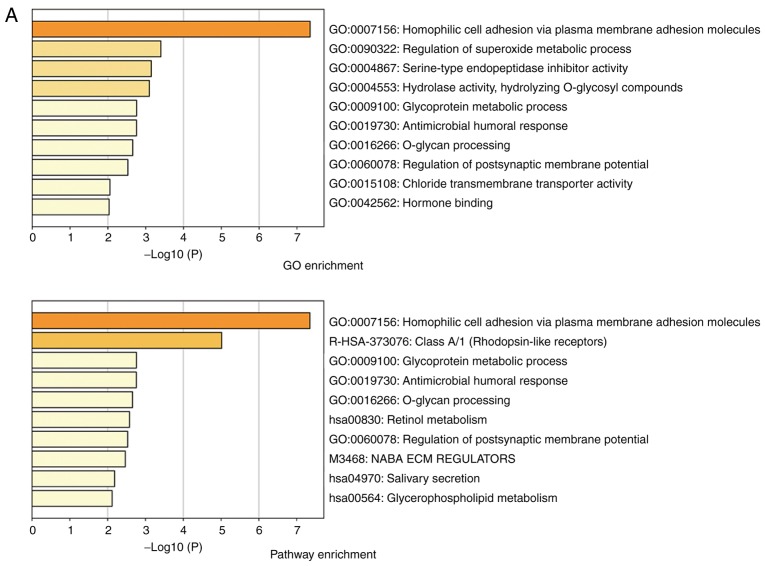
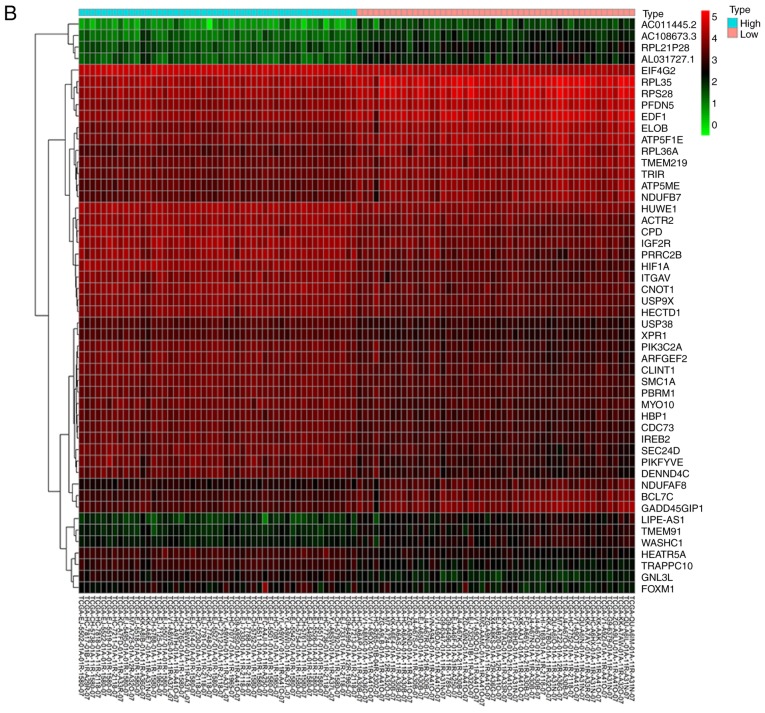
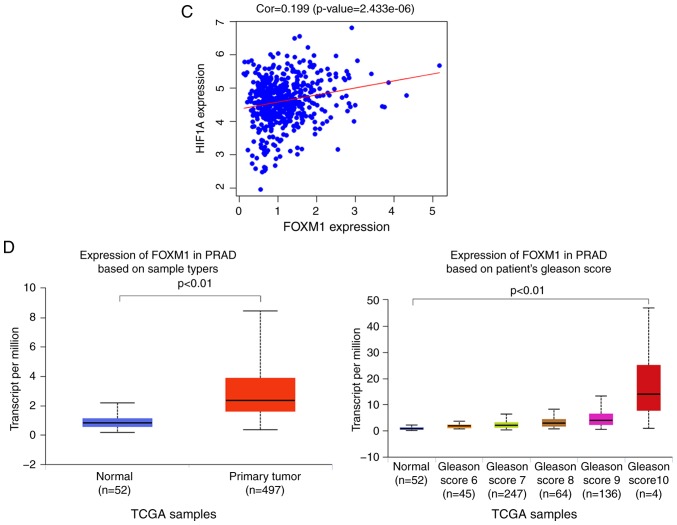
We next aimed to identify the possible molecular mechanisms that were responsible for hypoxia-induced EMT. GO enrichment analysis showed that high HIF-1α expression was associated with ‘homophilic cell adhesion via plasma membrane adhesion molecules’ (Fig. 2A). From the heatmap, we found that the expression of FoxM1 was upregulated in samples with high HIF-1α expression (Fig. 2B). Similar results were also obtained by correlation analysis, which showed that HIF-1α expression was positively correlated with FoxM1 expression (Fig. 2C). Furthermore, compared with normal tissue, upregulated expression of FoxM1 was detected in primary tumor samples (Fig. 2D); the expression of FoxM1 also increased gradually with higher Gleason scores (Fig. 2D). These results indicated that HIF-1α and FoxM1 may have important interactions, and their expression serves an important role in the invasion, migration and progression of PCa.
Figure 2.
Bioinformatics analysis and the correlation between HIF-1α and FoxM1 in PCa. (A) High and low expression of HIF1A TCGA samples were used to identify DEGs. The results of GO and pathway enrichment applied on top 200 upregulated DEGs are shown in the bar plot, the higher the column and the darker the color, the smaller the P-value, and the corresponding enrichment entry is marked on the right. (B) The relative expression difference of DEGs was shown in the heatmap, where the green and red represented low and high expression, respectively. DEGs, differentially expressed genes; GO, Gene Ontology; FoxM1, Forkhead box M1; HIF1A, hypoxia-inducible factor 1α; has, homo sapiens; PCa, prostate cancer; PRAD, prostate adenocarcinoma; TCGA, The Cancer Genome Atlas. (C) Pearson correlation analysis of HIF1A and FoxM1 in TCGA prostate samples was performed; the red line represented the expression trend of the two genes. (D) The expression of FoxM1 in PRAD and normal prostate tissues (left panel), and the relationship between FoxM1 and Gleason scores (right panel). DEGs, differentially expressed genes; GO, Gene Ontology; FoxM1, Forkhead box M1; HIF1A, hypoxia-inducible factor 1α; has, homo sapiens; PCa, prostate cancer; PRAD, prostate adenocarcinoma; TCGA, The Cancer Genome Atlas.
FoxM1 is required for hypoxia-induced EMT in PC3 and DU145 PCa cells
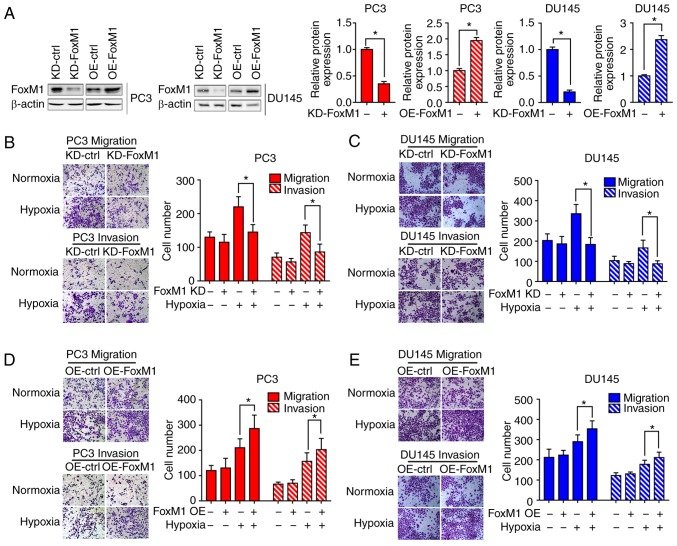
To determine whether hypoxia-induced EMT is dependent on FoxM1, FoxM1 knockdown (KD-FoxM1) and FoxM1 overexpression (OE-FoxM1) PC3 and DU145 (Fig. 3A) cell lines were established, and the expression was verified by western blot analysis. Different PC3 and DU145 cell clones were exposed to hypoxia or not for 48 h. Then, the cell migration (Fig. 3B and D) and invasive (Fig. 3C and E) abilities were determined by Transwell migration and invasion assays. Under hypoxic conditions, the number of cells migrating or invading through the chamber was significantly decreased in KD-FoxM1 groups compared with KD-control (ctrl) groups. In addition, the cell migration and invasive abilities were significantly increased in OE-FoxM1 than in OE-ctrl groups after exposure to hypoxia.
Figure 3.
FoxM1 mediates the hypoxia-induced metastatic phenotype of PC3 and DU145 cells. (A) Different PC3 and DU145 cell clones were established and analyzed by western blotting. (B and C) KD-ctrl and KD-FoxM1 PC3 and DU145 clones were pretreated under normoxic or hypoxic conditions for 24 h. The migration and invasion abilities were determined by Transwell migration and invasion assays. (D and E) OE-ctrl and OE-FoxM1 PC3 and DU145 clones were pretreated under normoxic or hypoxic conditions for 24 h. The migration and invasive abilities were determined by Transwell migration and invasion assays. Data are presented as the mean ± standard deviation of three replicates. *P<0.05. ctrl, control; FoxM1, Forkhead box M1; KD, knockdown; OE, overexpression.
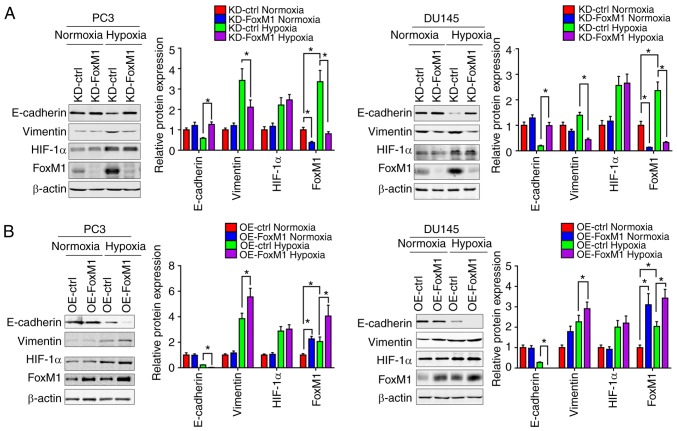
Furthermore, the protein levels of epithelial and mesenchymal phenotypic markers in PC3 and DU145 were measured by western blot analysis (Fig. 4A and B). Under hypoxic conditions, compared with the control groups, the expression of E-cadherin was increased in response to FoxM1 knockdown and decreased following FoxM1 overexpression, while the expression of vimentin was decreased by FoxM1 knockdown and increased by FoxM1 overexpression. Additionally, the expression of HIF-1α exhibited no significant change when the expression of FoxM1 was manipulated, suggesting that HIF-1α may not be regulated by FoxM1. The results indicated that FoxM1 serves an important role in the induction of EMT by hypoxia in PC3 and DU145 cells.
Figure 4.
FoxM1 mediates hypoxia-induced EMT in PC3 and DU145 cells. (A) KD-ctrl and KD-FoxM1 PC3 (left panel) and DU145 (right panel) clones were pretreated under normoxic or hypoxic conditions for 24 h. Then, the protein levels of EMT markers, E-cadherin and vimentin were detected by western blotting. (B) OE-ctrl and OE-FoxM1 PC3 (left panel) and DU145 (right panel) clones were pretreated under normoxic or hypoxic conditions for 24 h. Then, the protein levels were detected by western blotting. Data are presented as the mean ± standard deviation. of three replicates. *P<0.05. ctrl, control; FoxM1, Forkhead box M1; HIF1A, hypoxia-inducible factor 1α; KD, knockdown; OE, overexpression.
Inhibition of HIF-1α blocks the increase of FoxM1 induced by hypoxia at the mRNA level in PC3 and DU145 PCa cells
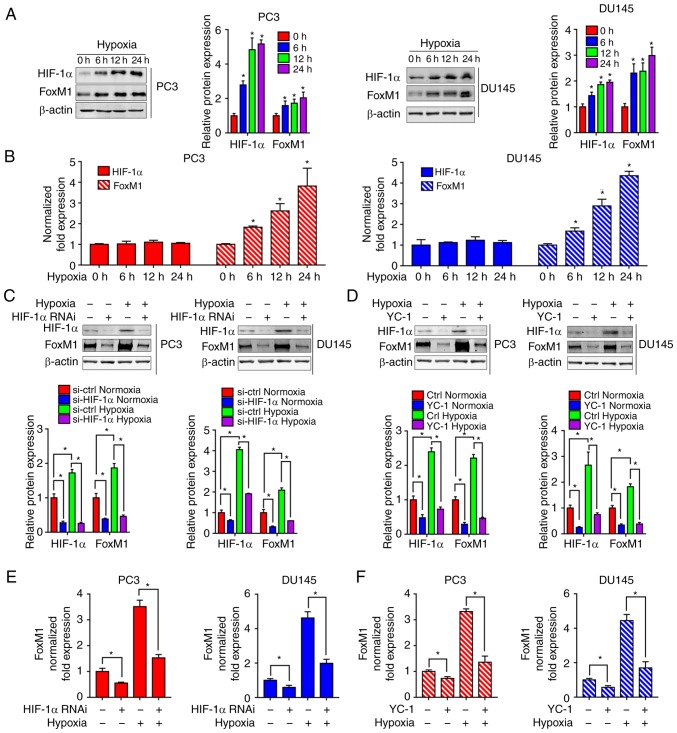
To further study the mechanism of hypoxia on FoxM1, the expression of HIF-1α and FoxM1 were evaluated by western blotting (Fig. 5A) and RT-qPCR (Fig. 5B) in PC3 and DU145 cells exposed to hypoxia for different durations (0, 6, 12 and 24 h). As presented in Fig. 5A and B, there were no significant changes in the expression of HIF-1α mRNA under different durations of hypoxia, while the protein expression levels of HIF-1α were significantly increased in hypoxia-exposed PC3 and DU145 cells compared with 0 h of exposure. Interestingly, increased FoxM1 expression was detected at the protein and mRNA levels, suggesting that FoxM1 was potentially regulated at the transcriptional level in PC3 and DU145 under hypoxia. As we proposed that HIF-1α was not regulated by FoxM1, we further investigated whether the induction of FoxM1 was regulated by HIF-1α. The RNA interference (Fig. 5C and E) and inhibitor YC-1 (Fig. 5D and 5F) of HIF-1α were used in PC3 and DU145 cells. The inhibitory effects of siRNA and YC-1 on HIF-1α expression was confirmed by western blotting (Fig. 5C and D). The results showed that knockdown of HIF-1α significantly inhibited hypoxia-induced overexpression of FoxM1 at the protein (Fig. 5C) and mRNA level (Fig. 5E). Similarly, HIF-1α inhibitor YC-1 was also found to significantly inhibit hypoxia-induced FoxM1 expression at the protein (Fig. 5D) and mRNA level (Fig. 5F).
Figure 5.
Inhibition of HIF-1α blocks hypoxia-induced upregulation of FoxM1. (A) PC3 (left panel) and DU145 (right panel) cells were cultured under normoxic or hypoxic conditions for different durations (0, 6, 12 and 24 h). HIF-1α and FoxM1 protein levels were detected by western blotting. (B) PC3 and DU145 cells were cultured under normoxic or hypoxic conditions for different durations (0, 6 12 and 24 h). HIF-1α and FoxM1 mRNA levels were detected by RT-qPCR. (C and D) PC3 and DU145 cells were transfected with specific RNAi duplexes targeting for HIF-1α or pretreated with HIF-1α inhibitor YC-1 for 24 h, and then exposed to hypoxia for an additional 24 h. Then, the protein expression levels of HIF-1α and FoxM1 were determined by western blotting. (E and F) Under similar conditions, the mRNA expression levels of FoxM1 were determined by RT-qPCR. Data are presented as the mean ± standard deviation of three replicates. *P<0.05. ctrl, control; FoxM1, Forkhead box M1; HIF-1α, hypoxia-inducible factor 1α; RNAi, RNA interference; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
HIF-1α regulates the transcription of FoxM1 during hypoxia in PC3 and DU145 PCa cells
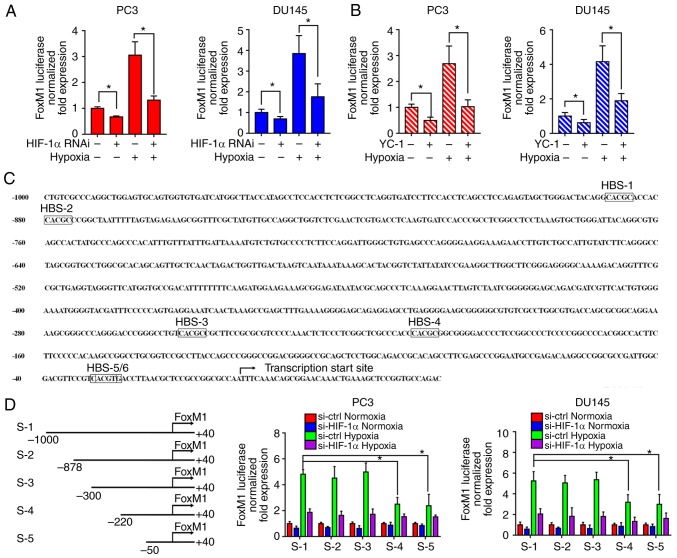
To investigate the response of the FoxM1 promoter to HIF-1α, RNA interference (Fig. 6A) and the inhibitor (Fig. 6B) of HIF-1α were applied to PC3 and DU145 cells. Then, FoxM1 promoter activities were evaluated by a dual-luciferase assay under normoxic or hypoxic conditions. The results showed that downregulation of HIF-1α significantly decreased the FoxM1 promotor activity under hypoxic conditions compared with the corresponding control. This suggested that HIF-1α activation is important for the transcriptional regulation of FoxM1. Furthermore, as shown in Fig. 6C, the promotor was investigated to identify potential HIF-1 binding sites. Six possible HIF-1 binding sites (HBS) were identified from upstream −1,000 bp to the translation start site: −885 bp (HBS-1, inverted), −875 bp (HBS-2, inverted), −249 bp (HBS-3, inverted), −203 bp (HBS-4, inverted), −25 bp (HBS-5, inverted), −23 bp (HBS-6, inverted). Therefore, different sequences of the FoxM1 promoter were cloned: S-1 (−1,000 bp ~ +40 bp), S-2 (−878 bp ~ +40 bp), S-3 (−300 bp ~ +40 bp), S-4 (−220 bp ~ +40 bp), S-5 (−50 bp ~ +40 bp) (Fig. 6D). The sequences were transfected into PC3 and DU145 cells then the transcriptional activities were detected by dual luciferase assay under normoxic or hypoxic conditions. Compared with the full length sequence S-1, sequences S-2 and S-3 did not reduce the transcription activity, while sequences S-4 and S-5 led to a significant decrease in activity. Additionally, the activities between S-4 and S-5 had no notable differences, while S-5 still exhibited transcriptional activity under hypoxia compared with the normoxic control. These results suggest HBS-3 and HBS-5/6, but not HBS-1, HBS-2 and HBS-4, are required for the FoxM1 transcriptional activity regulated by HIF-1α under hypoxic conditions.
Figure 6.
HIF-1α transcriptionally regulates FoxM1 under hypoxic conditions. (A and B) PC3 and DU145 cells were transfected with specific RNAi duplexes for HIF-1α or pretreated with HIF-1α inhibitor YC-1 for 24 h, and then exposed to hypoxia for additional 24 h. Then, the transcriptional activities of FoxM1 promotor were analyzed by luciferase assay under normoxic or hypoxic conditions. (C) Six possible HIF-1α binding sites were identified from upstream 1,000 to the translation start site of the FoxM1 promoter. The transcription start site was indicated. (D) Different lengths of reporter gene constructs (HBS-1 to 5, S-1 to 5) were designed. Then, the transcription activities of different constructs were analyzed by a dual-luciferase assay under normoxic or hypoxic conditions with or without HIF-1α knockdown in PC3 and DU145 cells. Data are presented as the mean ± standard deviation of three replicates. *P<0.05. ctrl, control; FoxM1, Forkhead box M1; HIF-1α, hypoxia-inducible factor 1α; HBS, HIF-1 binding sites; RNAi, RNA interference.
Discussion
EMT is a cell process through which cancer cells acquire increased metastatic potential (30). During this process, cells obtain a phenotype similar to fibroblasts and epithelial-specific protein markers, such as E-cadherin are downregulated, while that of mesenchymal protein markers such as vimentin are increased (31). The induction and regulation of EMT in PCa has been extensively studied by researchers. Accumulating evidence has suggested the association between hypoxia and EMT in PCa (32–34). We previously reported that overexpression of HIF-1α induced EMT in PCa LNCaP and PC3 cells both in vitro and in vivo (35). Mechanically, chronic hypoxia-induced slug promotes EMT of PCa cells by activating the expression of Ephrin-B1 (11). Additionally, Annexin A1 may be a key mediator of hypoxia-related EMT processes in PCa (36). Furthermore, monoamine oxidase A has been identified to induce EMT and stabilize HIF-1α, which then activates vascular endothelial growth factor (VEGF)-A/FOXO1/TWIST1 pathway in high-grade PCa (37). In our study, using bioinformatics analysis, we identified DEGs in PCa tissues with high or low expression of HIF-1α. Our data showed that DEGs which were associated with ‘homophilic cell adhesion via plasma adhesion molecules’ were of statistical significance between two groups exhibiting differential expression of HIF-1α. Changes in cell adhesion are considered as key elements in determining the development of invasive and metastatic tumors. Additionally, loss of cell adhesion is one of the hallmarks of EMT. In our study, EMT induction together with downregulation of E-Cadherin, which is a key cell adhesion molecule, were observed after exposure to hypoxia. However, the detail mechanisms involved in hypoxia induced-EMT in PCa remains unclear.
Recently, gene analysis using the high-throughput platforms has been developed as a promising tool with various clinical applications, such as the molecular diagnosis and classification of cancers, and the prediction of tumor response and patient prognosis. Several gene expression profiles related to PCa have been studied with microarray technology, revealing hundreds of DEGs that are involved in the process of tumorigenesis (38–40), serving a potential role in the identification of novel therapeutic targets. The present study applied bioinformatics analysis to identify DEGs in PCa with low or high expression of HIF-1α, with a particular focus on possible hub genes that are likely to play key roles in the progression of PCa. Under hypoxic conditions, cancer cells initiate a signaling pathway which triggers upregulation of the corresponding gene to adapt to the environment. These genes are regulated by the activation of the transcription factor HIF-1α, which serves an important role for the HIF family (41,42). In our study, several key DEGs were identified. Of those genes, FoxM1, which might be a potential target in other cancers was selected for further investigation.
FoxM1 is a transcription factor that belongs to the Forkhead superfamily and regulates the expression of target genes through the binding sequence TAAACA (43,44). FoxM1 has been reported to serve important roles in cell proliferation, cell cycle, cell differentiation, angiogenesis and metastasis (17,45). Previous studies have shown that in a variety of human malignant cancers FoxM1 is upregulated, which indicates the poor prognosis of patients (46–50). It was reported that FoxM1 expression in prostate epithelial cells is critical for prostate carcinogenesis (51). Additionally, the FoxM1 pathway was determined to act as a master regulator of PCa subtype 1 (PCS1) tumors, and targeting FoxM1 reduces cell growth and stemness in PCS1 tumors in vitro and in vivo (52). It was revealed that, in patients with PCa, high FoxM1 expression was associated with advanced tumor stages, high Gleason score and poor prognosis, suggesting the vital role of FoxM1 in PCa development and progression (53). Consistent with published data, in our study, the expression levels of FoxM1 were upregulated in PCa and were associated with Gleason scores as determined by bioinformatics analysis. Importantly, hypoxia-induced EMT was reported to be regulated by FoxM1. Our results imply that the inhibition of FoxM1 blocked the EMT process and FoxM1 is critical for hypoxia-induced EMT in PC3 and DU145 PCa cells.
In the present study, overexpression of exogenous FoxM1 or knockdown of FoxM1 alone had no notable effects on EMT and cancer cell migration/invasion. Although FoxM1 was identified to be a direct target and downstream of HIF-1α in PCa, the data we obtained suggested that FoxM1 might not directly mediate hypoxia/HIF-1α-induced EMT. There could be several reasons for this phenomenon. Firstly, under hypoxic conditions, the tumor microenvironment sustains major EMT-inducing pathways to facilitate tumor metastasis, such as transforming growth factor-β (TGFβ), nuclear factor-κB and Notch signaling pathways (54–56). Inflammatory cytokines including tumor necrosis factor α, TGFβ, interleukin (IL)-1, IL-6 and IL-8, secreted by surrounding inflammatory cells may also play an essential role in hypoxia-induced EMT (57–59). More importantly, there is growing experimental evidence that HIF-1α modulates the EMT by regulating the expression and activity of major transcription factors, including TWIST, SNAIL, SLUG, SIP1 and zinc finger E-box binding homeobox 1 (60). In our study, we failed to observed the direct effects FoxM1 on EMT in PCa. We speculated that hypoxia/HIF-1α-induced EMT in our model system may be mediated by an unknown transcriptional factor or a signaling pathway. Thus, we proposed the possible interplay between this unknown transcriptional factor and FoxM1. Based on published literature, hypoxia increases androgen receptor (AR) activity in PCa LNCaP cells (61) and androgens activate HIF-1, driving VEGF expression in androgen-sensitive LNCaP cells (62). Recently, a negative interplay/crosstalk between androgen/AR and hypoxia/HIF1 was proposed (63). Our unpublished data suggested that the transcription factors FoxM1, SOX9 and SOX2 are direct targets of AR. Given the fact that hypoxia/HIF1 was determined to associate with SOX9 (64) and SOX2 (65), we proposed that AR/SOX2 or AR/SOX9 signaling may be involved in crosstalk with FoxM1, leading to hypoxia/HIF1-induced EMT; however, further investigation is required. In addition, the particular genetic background of PCa may be another possible explanation. The results of our study may be PCa cell-type specific; further research is warranted to address this critical issue.
In our study, we also found that hypoxia upregulated the expression of HIF-1α at the protein level but not at mRNA level, suggesting the possible involvement of regulation of HIF-1α protein stability. However, the expression of FoxM1 increased significantly in a time-dependent manner at both the protein and mRNA levels under hypoxic condition. Previous studies have reported that HIF-1α could transcriptionally regulate FoxM1 in several cancer cell lines (66). To confirm this mechanism of hypoxia-induced EMT in PC3 and DU145 PCa cells, we used HIF-1α specific RNA interference to knockdown the expression and HIF-1α inhibitor YC-1 to inhibit the activity of HIF-1α. The results showed that reduction of HIF-1α significantly prevented FoxM1 protein and mRNA expression. The dual-luciferase assay also demonstrated that knockdown of HIF-1α expression reduced the activity of FoxM1 transcriptional promoter induced by hypoxia. In the process of HIF-1α regulating downstream target genes, the binding site on the target gene promoter which could bind HIF-1α is essential for transcriptional regulation (67,68). Therefore, the FoxM1 promoter was reconstructed and analyzed from upstream −1,000 to the translation start site. Our results demonstrated that the sequence from upstream −330 to the translation start site is critical for the transcriptional activation of FoxM1 regulated by HIF-1α. Analysis of different fragment length revealed that HBS-3 and HBS-5/6 on the FoxM1 promoter are likely to be binding sites for HIF-1α. These data indicated that FoxM1 is directly activated by HIF-1α binding to the HBS within the FoxM1 promoter during hypoxia-induced EMT.
In summary, our findings show that FoxM1 can be stimulated by hypoxia in PC3 and DU145 PCa cells which is dependent on the activation of the FoxM1 transcriptional promoter by HIF-1α. This induction of FoxM1 leads to the EMT of cells, involving decreased expression of epithelial protein markers, increased expression of mesenchymal protein markers and promoting cell invasive ability. This suggests that FoxM1 plays a key role in the hypoxia-induced EMT process of PCa. Future investigations are required to determine the suppression of FoxM1 as a potential therapeutic strategy for treating PCa.
Acknowledgements
Not applicable.
Funding
This study was partly supported by the National Natural Science Foundation of China (grant no. NSFC 81672538 to JZ).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CT, KW, SX and XW performed the experiments. CT and JZ wrote the manuscript. TL collected and interpreted the data and performed the bioinformatics analysis. KW, SX and XW provided technical assistance. DH and JZ designed this study and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Schatten H. Brief overview of prostate cancer statistics, grading, diagnosis and treatment strategies. Adv Exp Med Biol. 2018;1095:1–14. doi: 10.1007/978-3-319-95693-0_1. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Holm HV, Dahl AA, Klepp OH, Fossa SD. Modern treatment of metastatic prostate cancer. Tidsskr Nor Laegeforen. 2017;137:803–805. doi: 10.4045/tidsskr.16.0265. (In English, Norwegian) [DOI] [PubMed] [Google Scholar]
- 4.Grubb RL, III, Kibel AS. Prostate cancer: Screening, diagnosis and management in 2007. Mo Med. 2007;104:408–414. [PubMed] [Google Scholar]
- 5.Gomez CR, Kosari F, Munz JM, Schreiber CA, Knutson GJ, Ida CM, El Khattouti A, Karnes RJ, Cheville JC, Vasmatzis G, Vuk-Pavlović S. Prognostic value of discs large homolog 7 transcript levels in prostate cancer. PLoS One. 2013;8:e82833. doi: 10.1371/journal.pone.0082833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 8.Deep G, Panigrahi GK. Hypoxia-induced signaling promotes prostate cancer progression: Exosomes role as messenger of hypoxic response in tumor microenvironment. Crit Rev Oncog. 2015;20:419–434. doi: 10.1615/CritRevOncog.v20.i5-6.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du J, Sun B, Zhao X, Gu Q, Dong X, Mo J, Sun T, Wang J, Sun R, Liu Y. Hypoxia promotes vasculogenic mimicry formation by inducing epithelial-mesenchymal transition in ovarian carcinoma. Gynecol Oncol. 2014;133:575–583. doi: 10.1016/j.ygyno.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 10.Shaikh D, Zhou Q, Chen T, Ibe JC, Raj JU, Zhou G. cAMP-dependent protein kinase is essential for hypoxia- mediated epithelial-mesenchymal transition, migration, and invasion in lung cancer cells. Cell Signal. 2012;24:2396–2406. doi: 10.1016/j.cellsig.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, Liu Y, Yan X, Xu Y, Luo F, Ye J, Yan H, Yang X, Huang X, Zhang J, Ji G. HIFs enhance the migratory and neoplastic capacities of hepatocellular carcinoma cells by promoting EMT. Tumour Biol. 2014;35:8103–8114. doi: 10.1007/s13277-014-2056-0. [DOI] [PubMed] [Google Scholar]
- 12.Hay ED. Role of cell-matrix contacts in cell migration and epithelial-mesenchymal transformation. Cell Differ Dev. 1990;32:367–375. doi: 10.1016/0922-3371(90)90052-X. [DOI] [PubMed] [Google Scholar]
- 13.Khan MI, Hamid A, Adhami VM, Lall RK, Mukhtar H. Role of epithelial mesenchymal transition in prostate tumorigenesis. Curr Pharm Des. 2015;21:1240–1248. doi: 10.2174/1381612821666141211120326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowin P, Rowlands TM, Hatsell SJ. Cadherins and catenins in breast cancer. Curr Opin Cell Biol. 2005;17:499–508. doi: 10.1016/j.ceb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwasaki K, Ninomiya R, Shin T, Nomura T, Kajiwara T, Hijiya N, Moriyama M, Mimata H, Hamada F. Chronic hypoxia-induced slug promotes invasive behavior of prostate cancer cells by activating expression of ephrin-B1. Cancer Sci. 2018;109:3159–3170. doi: 10.1111/cas.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, Hu CJ, Bai JY. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16:57. doi: 10.1186/s12964-018-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S, Ali R, Sarkar FH. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010;122:337–346. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 19.Yang C, Chen H, Tan G, Gao W, Cheng L, Jiang X, Yu L, Tan Y. FOXM1 promotes the epithelial to mesenchymal transition by stimulating the transcription of Slug in human breast cancer. Cancer Lett. 2013;340:104–112. doi: 10.1016/j.canlet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Abdeljaoued S, Bettaieb I, Nasri M, Adouni O, Goucha A, El Amine O, Boussen H, Rahal K, Gamoudi A. Overexpression of FOXM1 is a potential prognostic marker in male breast cancer. Oncol Res Treat. 2017;40:167–172. doi: 10.1159/000458156. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Yu W, Zhou L, Wu S, Yang Y, Wang J, Tian Y, He D, Xu Y, Huang J, et al. Relationship among diet habit and lower urinary tract symptoms and sexual function in outpatient-based males with LUTS/BPH: A multiregional and cross-sectional study in China. BMJ Open. 2016;6:e010863. doi: 10.1136/bmjopen-2015-010863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong FF, Qu ZQ, Yuan HH, Wang JY, Zhao M, Guo YH, Shi J, Gong XD, Zhu YL, Liu F, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep. 2014;31:2660–2668. doi: 10.3892/or.2014.3129. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Kohashi K, Yamada Y, Maekawa A, Kuda M, Furue M, Oda Y. Prognostic significance of forkhead box M1 (FoxM1) expression and antitumour effect of FoxM1 inhibition in melanoma. Histopathology. 2016;69:63–71. doi: 10.1111/his.12909. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Liu Y, Yuan B, Yin L, Peng Y, Yu X, Zhou W, Gong Z, Liu J, He L, Li X. FOXM1 promotes the progression of prostate cancer by regulating PSA gene transcription. Oncotarget. 2017;8:17027–17037. doi: 10.18632/oncotarget.15224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, Wu D, Yu Q, Li L, Wu P. Prognostic value of FOXM1 in solid tumors: A systematic review and meta-analysis. Oncotarget. 2017;8:32298–32308. doi: 10.18632/oncotarget.15764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Method. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalluri RJ. EMT: When epithelial cells decide to become mesenchymal-like cells. J Clin Invest. 2009;119:1417–1419. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 32.Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 33.Boddy JL, Fox SB, Han C, Campo L, Turley H, Kanga S, Malone PR, Harris AL. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin Cancer Res. 2005;11:7658–7663. doi: 10.1158/1078-0432.CCR-05-0460. [DOI] [PubMed] [Google Scholar]
- 34.Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, Gouvin LM, Sharma VM, Mercurio AM. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: Implications for Gleason grading. Cancer Cell. 2010;17:319–332. doi: 10.1016/j.ccr.2010.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo Y, He DL, Ning L, Shen SL, Li L, Li X, Zhau HE, Chung LW. Over-expression of hypoxia-inducible factor-1alpha increases the invasive potency of LNCaP cells in vitro. BJU Int. 2006;98:1315–1319. doi: 10.1111/j.1464-410X.2006.06480.x. [DOI] [PubMed] [Google Scholar]
- 36.Bizzarro V, Belvedere R, Migliaro V, Romano E, Parente L, Petrella A. Hypoxia regulates ANXA1 expression to support prostate cancer cell invasion and aggressiveness. Cell Adh Migr. 2017;11:247–260. doi: 10.1080/19336918.2016.1259056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu JB, Shao C, Li X, Li Q, Hu P, Shi C, Li Y, Chen YT, Yin F, Liao CP, et al. Monoamine oxidase A mediates prostate tumorigenesis and cancer metastasis. J Clin Invest. 2014;124:2891–2908. doi: 10.1172/JCI70982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheng WS, Tao H, Hu EP, Liu S, Cai HR, Tao XL, Zhang L, Mao JJ, Yan DL. Both genes and lncRNAs can be used as biomarkers of prostate cancer by using high throughput sequencing data. Eur Rev Med Pharmacol Sci. 2014;18:3504–3510. [PubMed] [Google Scholar]
- 39.Han Y, Jin X, Li H, Wang K, Gao J, Song L, Lv Y. Microarray analysis of copy-number variations and gene expression profiles in prostate cancer. Medicine (Baltimore) 2017;96:e7264. doi: 10.1097/MD.0000000000007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan J, Jin X, Wang K. Integrated bioinformatics analysis of potential biomarkers for prostate cancer. 2019;25:455–460. doi: 10.1007/s12253-017-0346-8. [DOI] [PubMed] [Google Scholar]
- 41.Soni S, Padwad YS. HIF-1 in cancer therapy: Two decade long story of a transcription factor. 2017;56:503–515. doi: 10.1080/0284186X.2017.1301680. [DOI] [PubMed] [Google Scholar]
- 42.Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 43.Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG, Clevers H. The human TRIDENT/HFH- 11/FKHL16 gene: Structure, localization, and promoter characterization. Genomics. 1997;46:435–442. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- 44.Laoukili J, Stahl M, Medema RH. FoxM1: At the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52:74–84. doi: 10.1016/j.semcancer.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 46.Yang DK, Son CH, Lee SK, Choi PJ, Lee KE, Roh MS. Forkhead box M1 expression in pulmonary squamous cell carcinoma: Correlation with clinicopathologic features and its prognostic significance. Hum Pathol. 2009;40:464–470. doi: 10.1016/j.humpath.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Sun HC, Li M, Lu JL, Yan DW, Zhou CZ, Fan JW, Qin XB, Tang HM, Peng ZH. Overexpression of Forkhead box M1 protein associates with aggressive tumor features and poor prognosis of hepatocellular carcinoma. Oncol Rep. 2011;25:1533–1539. doi: 10.3892/or.2011.1230. [DOI] [PubMed] [Google Scholar]
- 48.Huynh KM, Soh JW, Dash R, Sarkar D, Fisher PB, Kang D. FOXM1 expression mediates growth suppression during terminal differentiation of HO-1 human metastatic melanoma cells. J Cell Physiol. 2011;226:194–204. doi: 10.1002/jcp.22326. [DOI] [PubMed] [Google Scholar]
- 49.Xia L, Mo P, Huang W, Zhang L, Wang Y, Zhu H, Tian D, Liu J, Chen Z, Zhang Y, et al. The TNF-α/ROS/HIF-1-induced upregulation of FoxMI expression promotes HCC proliferation and resistance to apoptosis. Carcinogenesis. 2012;33:2250–2259. doi: 10.1093/carcin/bgs249. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Wen L, Zhao SH, Ai ZH, Guo JZ, Liu WC. FoxM1 expression is significantly associated with cisplatin-based chemotherapy resistance and poor prognosis in advanced non-small cell lung cancer patients. Lung Cancer. 2013;79:173–179. doi: 10.1016/j.lungcan.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V, Zhang Y, Paluch AM, Waltz SE, Kasper S, Kalin TV. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J Biol Chem. 2013;288:22527–22541. doi: 10.1074/jbc.M113.455089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ketola K, Munuganti RSN, Davies A, Nip KM, Bishop JL, Zoubeidi A. Targeting prostate cancer subtype 1 by forkhead box M1 pathway inhibition. Clin Cancer Res. 2017;23:6923–6933. doi: 10.1158/1078-0432.CCR-17-0901. [DOI] [PubMed] [Google Scholar]
- 53.Kim MY, Jung AR, Kim GE, Yang J, Ha US, Hong SH, Choi YJ, Moon MH, Kim SW, Lee JY, Park YH. High FOXM1 expression is a prognostic marker for poor clinical outcomes in prostate cancer. J Cancer. 2019;10:749–756. doi: 10.7150/jca.28099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 55.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannoni E, Bianchini F, Calorini L, Chiarugi P. Cancer associated fibroblasts exploit reactive oxygen species through a proinflammatory signature leading to epithelial mesenchymal transition and stemness. Antioxid Redox Signal. 2011;14:2361–2371. doi: 10.1089/ars.2010.3727. [DOI] [PubMed] [Google Scholar]
- 57.Chuang MJ, Sun KH, Tang SJ, Deng MW, Wu YH, Sung JS, Cha TL, Sun GH. Tumor-derived tumor necrosis factor-alpha promotes progression and epithelial-mesenchymal transition in renal cell carcinoma cells. Cancer Sci. 2008;99:905–913. doi: 10.1111/j.1349-7006.2008.00756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, Oberyszyn TM, Hall BM. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28:2940–2947. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.St John MA, Dohadwala M, Luo J, Wang G, Lee G, Shih H, Heinrich E, Krysan K, Walser T, Hazra S, Zhu L, et al. Proinflammatory mediators upregulate snail in head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:6018–6027. doi: 10.1158/1078-0432.CCR-09-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang J, Tang YL, Liang XH. EMT: A new vision of hypoxia promoting cancer progression. Cancer Biol Ther. 2011;11:714–723. doi: 10.4161/cbt.11.8.15274. [DOI] [PubMed] [Google Scholar]
- 61.Park SY, Kim YJ, Gao AC, Mohler JL, Onate SA, Hidalgo AA, Ip C, Park EM, Yoon SY, Park YM. Hypoxia increases androgen receptor activity in prostate cancer cells. Cancer Res. 2006;66:5121–5129. doi: 10.1158/0008-5472.CAN-05-1341. [DOI] [PubMed] [Google Scholar]
- 62.Mabjeesh NJ, Willard MT, Frederickson CE, Zhong H, Simons JW. Androgens stimulate hypoxia-inducible factor 1 activation via autocrine loop of tyrosine kinase receptor/phosphatidylinositol 3′-kinase/protein kinase B in prostate cancer cells. Clin Cancer Res. 2003;9:2416–2425. [PubMed] [Google Scholar]
- 63.Geng H, Xue C, Mendonca J, Sun XX, Liu Q, Reardon PN, Chen Y, Qian K, Hua V, Chen A, et al. Interplay between hypoxia and androgen controls a metabolic switch conferring resistance to androgen/AR-targeted therapy. Nat Commun. 2018;9:4972. doi: 10.1038/s41467-018-07411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amarilio R, Viukov SV, Sharir A, Eshkar-Oren I, Johnson RS, Zelzer E. HIF1alpha regulation of Sox9 is necessary to maintain differentiation of hypoxic prechondrogenic cells during early skeletogenesis. Development. 2007;134:3917–3928. doi: 10.1242/dev.008441. [DOI] [PubMed] [Google Scholar]
- 65.Bae KM, Dai Y, Vieweg J, Siemann DW. Hypoxia regulates SOX2 expression to promote prostate cancer cell invasion and sphere formation. Am J Cancer Res. 2016;6:1078–1088. [PMC free article] [PubMed] [Google Scholar]
- 66.Xia LM, Huang WJ, Wang B, Liu M, Zhang Q, Yan W, Zhu Q, Luo M, Zhou ZZ, Tian DA. Transcriptional up-regulation of FoxM1 in response to hypoxia is mediated by HIF-1. J Cell Biochem. 2009;106:247–256. doi: 10.1002/jcb.21996. [DOI] [PubMed] [Google Scholar]
- 67.Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- 68.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.