Abstract
Background
Polyphenol catechins from green tea, particularly (−)-epigallocatechin-3-gallate (EGCG), exhibits numerous beneficial health effects, although the mechanisms remain unclear.
Methods
In this study, the mechanism of EGCG-mediated healing in an experimentally injured zebrafish model was examined at the cellular and molecular level using confocal microscopy and gene expression analysis.
Results
The mechanisms of action of EGCG were shown to involve: (1) reducing neutrophil response (accumulation, travel speed, and distance) and (2) downregulating the expression of IL-1β, TNFα, and related signaling pathways. As determined by dynamic time-lapse tracking studies, the local accumulation of neutrophils with high migration speeds after wounding (n=33 cells, v=0.020 μm/s, d=37.8 μm), underwent significant reduction following treatment with EGCG doses of 300 μM (n=22 cells, v=0.013 μm/s, d=39.5 μm) and 600 μM (n=18 cells, v=0.008 μm/s, d=9.53 μm). Reverse transcription polymerase chain reaction studies revealed that several signature genes in the IL-1β, TNFα, and related signaling pathways were downregulated after EGCG treatment.
Conclusion
The convenience, transparency, and simplicity of the zebrafish model facilitate tracking of fluorescent neutrophils in real time, in order to monitor inflammation, and assess the impact of therapeutic agents.
Keywords: green tea, innate immunity, animal models, IL-1, TNF
Introduction
Acute inflammation is an important early immune response that protects the host against injury and pathogen invasion.1 Inflammation is initiated when activated stromal cells and other innate immune cells secrete specific mediators and cytokines or chemokines into surrounding extracellular space, thus promoting the recruitment of leukocytes to stressed tissues.2 Pro-inflammatory neutrophils are rapidly recruited after infection, and are aggressive in removing invading microorganisms. Fifty or more pro-inflammatory molecules are localized on the plasma membrane of neutrophils and within their intracellular granules, including oxygen metabolites, proteolytic enzymes, and other chemical mediators that effectively destroy any living tissues with which they come in contact.3 Neutrophils also release type I interferon and cyclin-dependent kinase to sustain inflammation.1,4,5 Under normal conditions, neutrophils also promote wound healing. After eliminating target organisms, activated neutrophils generate anti-inflammatory agents such as lipoxin, macrophage-derived resolvins, and protectins, and secretory leukocyte protease inhibitors to attenuate further leukocyte infiltration and accumulation.6–8 Interventions targeting neutrophils are being developed to complement treatments for autoinflammatory and autoimmune conditions.9,10
Conventional studies involving neutrophils require isolating the cells under natural conditions or indirectly studying the cells by examining their excreted metabolites, although advances in genetic modulation techniques now enable both steps to be conducted in a single model organism, such as the zebrafish, Danio rerio. Traditionally, mice have been the most popular animal model in immunology research. However, the costs associated with facilities for maintaining mice and conducting large-scale projects are typically high, and this can negatively impact efforts to meet the high demand for testing new drugs. Zebrafish possess several attributes that make an attractive candidate in research, including their rapid growth rate, high fecundity, and versatility in image analyses. Optical transparency, which enables direct visualization of biological processes, has made zebrafish a particularly useful model for studying embryonic development and organogenesis since the 1980s.11 The availability of clones of homozygous fish and ease of breeding have made zebrafish a promising tool for vertebrate genetic analysis.12 Genetic studies conducted using gamma-irradiated fish mutants have identified genes associated with human disorders.13–16 Recent interest has also focused on utilizing zebrafish in high throughput screening of therapeutic compounds. For example, the weiẞherbst zebrafish mutant develops hypochromic anemia and is thus useful as a disease model for drug screening.15,16
Indeed, zebrafish provide a versatile model for studying immune responses. As they are exposed to pathogens at an early age, most fish larvae possess a highly developed innate immune system. The innate immune system develops as soon as 2 days post-fertilization, and the adaptive immune system is functional by 4–6 weeks post-fertilization.17 Since thymic organogenesis and compartmentalization into the medulla and cortex are well conserved in zebrafish,18 the immune system is “fully armed” with cells of major hematopoietic lineages (erythrocyte, leukocytes, myeloid cells). Genome-wide mutagenesis screening studies revealed that hematopoiesis and lymphopoiesis in zebrafish are similar to the processes in the human bone marrow.19,20 A library of 24 toll-like receptors has been described in zebrafish, including 10 orthologs of human toll-like receptor family members.21,22
Reporter fish with fluorescent-labeled immune cells have been developed to track macrophages, neutrophils, and epithelial cells.23 Since studying neutrophils could open up opportunities for identifying drugs with anti-inflammatory effects, many studies have investigated the use of mpx mutants with GFP-labeled neutrophils as a testing platform. One popular experimental design involves simple tail resection to induce inflammation and monitoring of real-time trafficking of neutrophils inside living, translucent fish.24 Based on their high fecundity and translucent vasculature, zebrafish represent a straightforward, cost-effective model organism for testing chemical compounds and rapidly assessing responses at the cellular level.
There is considerable interest in using natural compounds for treating chronic and acute inflammation. Green tea is a traditional drink known for its anti-inflammatory effects. The major catechin in green tea, (−)-epigallocatechin-3-gallate (EGCG), accounts for most of the anti-inflammatory activity.25 EGCG suppresses multiple pro-inflammatory pathways, including the NF-κB pathway and allergic IgE-mediated histamine release.26,27 The anti-inflammatory properties of EGCG have been demonstrated by its effects on reducing trans-endothelial migration of cultured neutrophils and inhibiting the production of reactive oxygen species.28,29 In the present study, we demonstrate the utility of transgenic zebrafish as a suitable model for anti-inflammatory drug testing using EGCG as a modulator.
Materials and methods
Zebrafish husbandry
Tg(mpx:gfp) zebrafish were obtained from Zebrafish International Resource Center, Eugene, Oregon, and maintained on a 14 hr/10 hr light/dark cycle at 28.5°C in a zebrafish facility (Center for Nuclear Receptor and Cell Signaling, University of Houston, Houston, TX) equipped with Tecniplast (West Chester, PA) tank system. After natural spawning, embryos were grown in a 28°C incubator with daily renewal of fish medium (E3) and removal of spoiled embryos until desired maturation was reached for experimentation. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (NAP 2011) and the animal use protocol was approved by the Institutional Animal Care and Use Committee at the University of Houston.
Tail resection, EGCG treatment
At 4 days post fertilization (dpf), Tg(mpx:gfp) larvae were anaesthetized in approximately 200 µg/mL (0.02%) MS-222 (Millipore Sigma, Burlington, MA, USA)-infused E3. All media were sterile, and razor blades were cleaned with 70% ethanol before use. Tail-fin resection was performed using a sterile disposable razor blade at the distal-most point of the pigment gap. Fish recovered in 12-well plates with 10 fish per well and 2 mL of fresh medium infused with 300 μM or 600 μM EGCG (Millipore Sigma, Burlington, MA, USA).
Short-time-frame imaging
After 4–5 hrs, a total of 3–4 of untreated or treated, anesthetized larvae were mounted laterally on a glass slide and imaged using AxioObserver D.1 epifluorescence microscope (Carl Zeiss, Thornwood, NY, USA). For quantification of neutrophil accumulation, manual cell counts and intensity measurements were performed. For manual counting, two researchers were randomly assigned to count the number of cells in the region extending up to 100 μm from the transection site. For intensity measurements, the total intensity in the same spot was measured using Fiji ImageJ (NIH, Madison, WI, USA) with correction for background signal. The following formula was used for quantification: adjusted total fluorescence = integrated density – (area of selected cell × mean background fluorescence). The data were fed into GraphPad Prism (GraphPad, San Diego, CA, USA) for analysis by unpaired Student's t-test and one-way ANOVA, with p-values of 0.001, 0.01, and 0.05 considered significant. Graphs show mean values ± SEM for 10 fish.
Prolonged time-lapse imaging
Low-melt agarose (1%) (Bio-Rad, Hercules, CA, USA) was brought to liquid state at 40°C and infused with EGCG to a final concentration of 300 or 600 μM. Anesthetized larvae were quickly mounted alive in infused low-melt agarose in a six-well glass-bottom plate (MatTek, Ashland, MA, USA). Time-lapse images were acquired using super-resolution Olympus FV1000 multiphoton confocal laser scanning microscope (Olympus, Center Valley, PA, USA) equipped with a 20× subjective, at a time span of 1 min over at least 200 mins after tail-fin resection. Migration velocity and distance traveled were calculated and plotted using the manual tracking plug-in of Fiji ImageJ.
Gene expression
A total of nine untreated and EGCG-treated 4 dpf larvae were anesthetized and RNA isolated according to SingleShot mRNA kit instructions (Bio-Rad, Hercules, CA, USA). RNA was pooled from three fish for each condition, to obtain sufficient RNA. The concentration of isolated mRNA was checked using a DS-11 spectrophotometer (DeNovix, Wilmington, DE, USA). cDNA was synthesized using iScript (Bio-Rad, Hercules, CA, USA) following manufacturer instructions, followed by qPCR in 10 µL total volume of SsoAdvanced Universal SYBR Green supermix, using 200 ng cDNA for each gene. Primers for 34 zebrafish-specific inflammation-related genes (Table 1) were designed and pre-spotted in 96-well PrimePCR plates (Bio-Rad, Hercules, CA, USA). qPCR was performed in CFX96 cycler under PrimePCR setting: initialized as 10 mins-95°C, followed by 40 cycles 15 s–95°C, 1 min–60°C, with a melt curve cycle. Relative expression was calculated and normalized against three housekeeping genes: actin beta 1 actb1, elongation factor 1 elf1, and tubulin alpha tuba.
Table 1.
Genes interrogated by RTPCR, modulated by tail resection and EGCG treatment
| Gene change after | Gene change after | T-test | |
|---|---|---|---|
| Tail resectiona | EGCG-treatmentb | ||
| Gene | Mean ± SE (%) | Mean ± SE (%) | p-Value |
| ATF2 | 69.1±6.5% | 37.8±2.5% | 0.18 |
| CHUK | 128.5±12.5% | 85.8±6.0% | 0.31 |
| CP | 147.1±31.1% | 154.9±25.0% | 0.42 |
| F3A | 101.9±21.4% | 281.3±45.2% | 0.23 |
| HMOX1 | 375.6±82.5% | 531.1±81.0% | 0.66 |
| IL-8A | 125.8±7.6% | 112.2±9.3% | 0.66 |
| IL1B | 233.4±15.0% | 49.5±4.5% | 0.03 |
| IL1RAPL1A | 144.9±31.5% | 78.7±10.1% | 0.47 |
| IRF1A | 87.8±20.0% | 40.3±5.8% | 0.43 |
| M17 | 145.8±11.5% | 73.9±3.2% | 0.11 |
| NFKB1 | 125.0±8.1% | 108.3±3.7% | 0.50 |
| NFKBIB | 94.3±4.8% | 72.8±2.6% | 0.22 |
| PTGS2A | 232.5±19.4% | 141.7±22.2% | 0.27 |
| SERPINE1 | 257.9±84.5% | 83.0±5.8% | 0.48 |
| TNFB | 127.4±4.5% | 81.8±4.9% | 0.04 |
| TRAF6 | 410.8±113.0% | 131.2±37.6% | 0.41 |
| UBB | 129.1±11.0% | 82.4±2.0% | 0.21 |
Notes: aThe percent expression level after tail resection represents mean ± SE of n=3 replicates (each representing three fish), with the level in control fish (no tail resection) being normalized to 100%. bThe percent expression level after EGCG treatment represents mean ± SE of n=3 replicates (each representing three fish), with the level in control fish (no tail resection) being normalized to 100%. The corresponding t-test p-values are listed in the last column. Significant changes are signified by bold.
Abbreviation: EGCG, epigallocatechin-3-gallate.
Statistics
Data were analyzed and plotted using GraphPad Prism 5. Kolmogorov–Smirnov test was used to assess the normality of the data. For comparisons of multiple groups, ANOVA test and subsequent post-test pairwise comparisons were used, employing the unpaired t-test, with p-values <0.05 considered significant. Preliminary treatment studies revealed ~34 neutrophils in control fish, and 20 neutrophils in treated fish, with a standard deviation of 6. Power calculations reveal that even a sample size of 3 per group would be sufficient to detect differences between groups at 0.05 level of significance, with 80% power. Hence, 3–10 fish were included in all studies.
Results
Static quantification of neutrophil recruitment demonstrates suppressive impact of EGCG
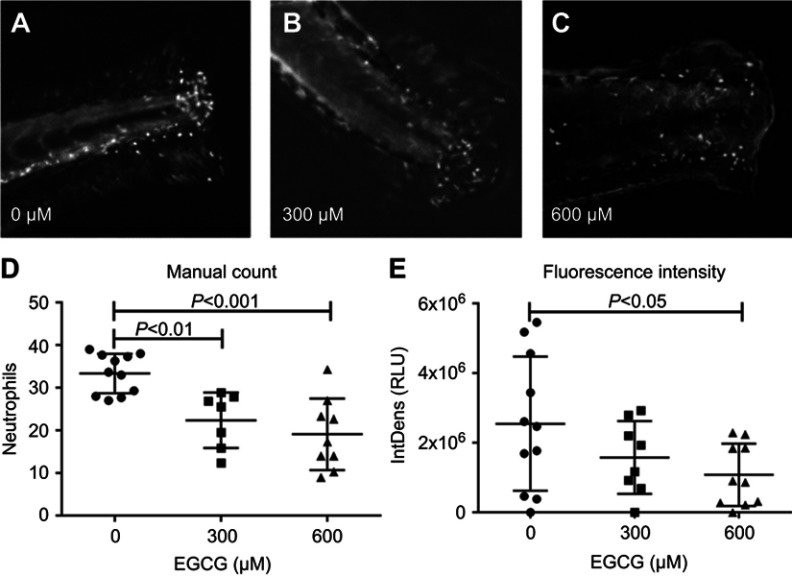
Tg(mpx:gfp) zebrafish are useful as a model of inflammation because they allow the tracking of GFP-labeled neutrophils within the transparent fish.24 Using this model, the process of inflammation and anti-inflammatory effects of EGCGs was captured in real time. In brief, 4 dpf larvae were anesthetized for tailfin-resection and mounted live for tracking neutrophil responses. Animated recordings of neutrophil migration or accumulation were obtained using an inverted fluorescence confocal microscope for up to 4 hrs after tail resection. Untreated fish exhibited an increased number of neutrophils at the injury site compared to fish treated with 300 or 600 μM EGCG (Figure 1A–C). At the 4 hrs mark, we found the highest number of neutrophils in untreated fish, with an average of 33 cells (Figure 1D), and this number was significantly lower in fish exposed to 300 μM EGCG (n=22 cells, p=0.003) or 600 μM EGCG (n=18 cells, p=0.0006). As the neutrophils were fluorescent, we confirmed these findings by quantifying the fluorescence intensity at the local injury site using automated ImageJ software. The intensity declined from 2.8×106 LU in untreated fish to 1.8×106 LU (p=0.17) and 1.2×106 LU (p=0.04) in fish treated with 300 or 600 μM EGCG (Figure 1E). EGCG at doses >900 μM were lethal to the larvae, even with short exposure; hence, the maximum dose used in this study was 600 μM.
Figure 1.
EGCG reduced neutrophil accumulation in tail-resected larvae. (A–C) 4-dpf zebrafish were tail-resected and incubated with EGCG for 4 hrs. Fluorescent neutrophils were imaged within the region of interest (ROI) (5 mm from resected line). Overall, more neutrophil accumulation events were observed in untreated control zebrafish compared to EGCG-treated fish (A–C). Plotted in (D) are the number of neutrophils and plotted in (E) is the fluorescence intensity observed in one ROI. The data show a consistent dose-dependent effect of EGCG. On average, 33 neutrophils were recruited to the resection site without EGCG, but this dropped to 22 and 18 cells, or from 2.8×106 light units (LU) to 1.8 and 1.2×106 LU, in fish treated with 300 or 600 μM EGCG (mean ± SD, n=10, t-test).
Abbreviation: EGCG, epigallocatechin-3-gallate.
EGCG reduces the frequency of neutrophil migration and dynamic motion following injury
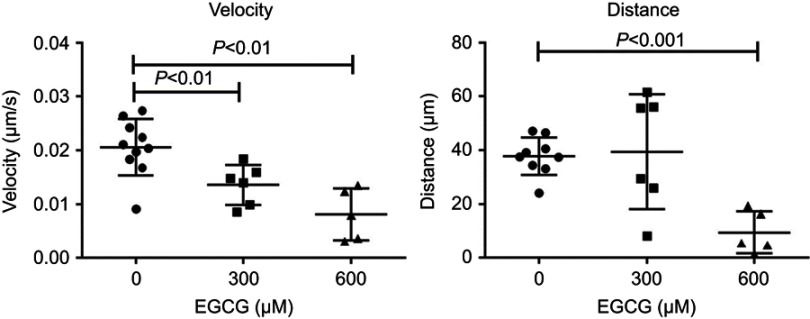
For time-lapse tracking analyses, neutrophil migration to the injury site was monitored after tail resection, with or without EGCG treatment. Neutrophils were tracked over the first 2 hrs post-injury, with or without EGCG, using confocal fluorescence microscopy, and the data were analyzed using Fiji Manual Tracking. All neutrophils in the untreated group (d=37.8 μm) traveled a greater distance than cells from fish treated with 600 μM EGCG (d=9.53 μm, p=0.0002), but a similar distance compared with neutrophils from fish treated with 300 μM EGCG (d=39.5 μm, ns) (Figure 2). The average velocity of neutrophil migration in the untreated fish (v=0.020 μm/s) was significantly greater than that in fish treated with EGCG at 300 μM (v=0.013 μm/s, p=0.008) or 600 μM (v=0.008 μm/s, p=0.001), as shown in Figure 2. EGCG treatment reduced leukocyte infiltration in terms of recorded tracks, average speed, and mean distance traveled by these cells, as illustrated in Figure 3. The attendant movie illustrates the dynamic attenuation of the neutrophil recruitment associated with EGCG treatment, fixed frames of which are shown in Figure 3.
Figure 2.
EGCG suppressed neutrophil migration speed and traveled distance. The larvae at 4 dpf larvae were tail-resected and mounted in EGCG infused agar for 2 hr imaging. From the time-lapse records, neutrophils were detected by fluorescence microscopy. Motion parameters including velocity and distance were extracted using Fiji Manual Tracking. More dynamic tracks were recorded in the untreated control fish within the ROI, compared to the ECGC treated fish, in terms of the number of active tracks, distance, and speed. Quantitative analysis of event speed (LEFT) and distance (RIGHT) in ROI of zebrafish treated with vehicle and EGCG (300 and 600 μM) are plotted (mean ± SD, n=6–10, t-test).
Abbreviations: EGCG, epigallocatechin-3-gallate; ROI, region of interest.
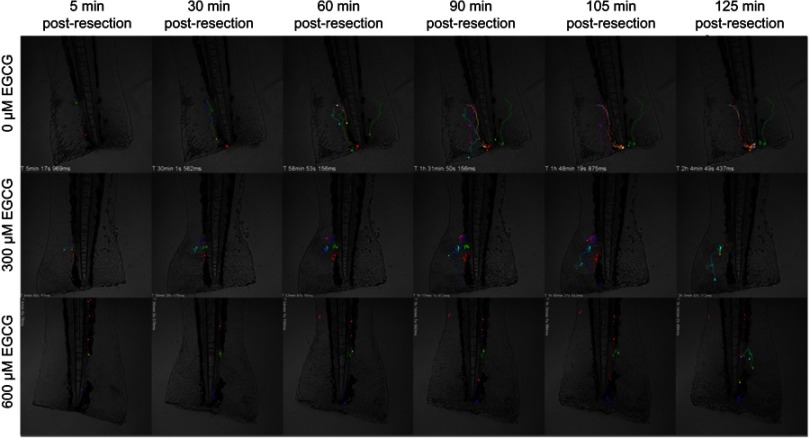
Figure 3.
EGCG suppressed neutrophil movement toward the inflamed region. Extracted frames from time-lapse fluorescence tracking records showed less defined tracks in fish treated with EGCG. A dynamic view of these events can be seen in the attached movie (Supplementary animations).
Abbreviation: EGCG, epigallocatechin-3-gallate.
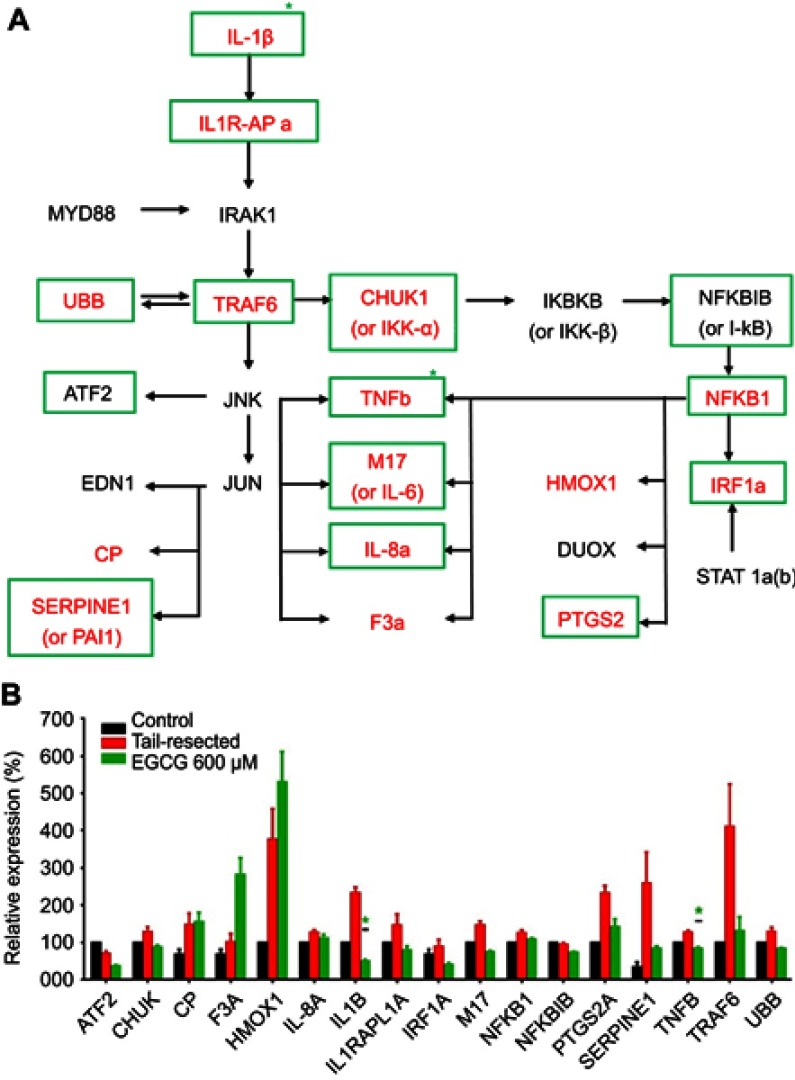
Identifying EGCG targets in inflammation signaling pathways, in tail-fin resected zebrafish
Based on the published data on how EGCG impacts various pathways, we examined a select set of 34 genes implicated in inflammation, including IL-1 and NF-kB driven pathways. Following tail resection, 15 out of the 34 genes interrogated were elevated at least 10%, compared to fish with intact tails (Table 1); these are presented in Figure 4A in red bold font. Next, we asked which genes were downregulated by EGCG. Of the tail resection induced genes, 12 genes were downregulated at least 10% following EGCG treatment, as indicated by green boxes in Figure 4. Of these EGCG-induced changes, two reductions attained statistical significance, namely IL1B (p=0.026) and TNFB (p=0.045). Overall, as evident from Figure 4, several inflammatory cytokines (eg, IL-1β, TNFβ, IL-6) and inflammation related signaling molecules (eg, IL1R-APa, TRAF6, IRF1a, etc.) showed a trend toward upregulation following tail resection, but reduction following EGCG administration, though most of these changes failed to attain statistical significance.
Figure 4.
Impact of EGCG on wounding-induced genes in the interleukin 1β and related signaling pathway. A group of nine tail-fin resected zebrafish were exposed to EGCG for 4 hrs. mRNA was collected and pooled (three fish RNA per pool) for RTPCR amplification of a library of 34 candidate genes in the interleukin-1 beta and related signaling pathway, as detailed in Table 1. The pathway diagram shown in (A) represents likely gene interaction relationships between the interrogated genes, constructed based on the literature review. Only genes that exhibited at least 10% change compared to the control group are plotted. The RNAs corresponding to the molecules in red-bold letters (in “a”) were upregulated following tail-fin resection by at least 10%, compared to the control fish. The RNAs corresponding to the molecules within green boxes were downregulated at least 10% after EGCG treatment (in “a”). Plotted in (B) are the mean (+ standard error) gene expression changes in three replicates (three fish per replicate), with all data normalized to the expression levels in control un-resected, untreated fish. Green asterisks mark significant fold-changes. In tail-resected zebrafish, wounding led to the upregulation of 15 inflammatory genes, of which 11 were subdued following EGCG treatment, of which IL1B and TNFB attained statistical significance.
Abbreviations: EGCG, epigallocatechin-3-gallate; RTPCR, reverse transcription pollymerase chain reaction.
Discussion
Neutrophils are among the earliest immune cells recruited in response to tissue injury. The zebrafish model provides researchers unique opportunities to visualize neutrophil migration in real time. In this study, we examined the protective effects of EGCG against inflammation in tailfin-resected model of transgenic zebrafish. In our study, we observed a sizable average number of neutrophils (an average of 33 cells) being recruited to the wound site, which was indicative of a normal inflammatory response to injury. Because previous reports documented that neutrophil recruitment persists for approximately 8 hrs after wounding, with a peak at 4 hrs, we examined neutrophil recruitment at 4 hrs after resection.24 Our data indicate reductions from an average of 33 cells in untreated fish to an average of 22 and 18 cells in fish treated with EGCG at 300 or 600 μM (~31% and 43% reduction), respectively. Live recording of neutrophil recruitment also clearly demonstrated the delay in the recruitment of neutrophils. The frequency of qualified neutrophil migration tracks in untreated fish was twice as high as that seen in EGCG-treated fish. The speed and distance traveled were also lower in EGCG-treated fish. These findings are consistent with previous reports of the inhibitory effect of EGCG on neutrophil function and mobility.28,29
Neutrophils are recruited by signals released from damaged and necrotic cells. The process of neutrophil extravasation from the vasculature into the tissues is governed by chemokines and cytokines, including IL-1β,30,31,33, CXCL8,32 hydrogen peroxide,23 and lipid mediators. It is also well known that neutrophils are capable of processing and producing active IL-1β. EGCG dampens multiple innate signaling pathways including histamine release, 27 reactive oxygen species production,28 and production of chemoattractants such as CINC1, IL-8,29 TNF-α25,34, and IL-1β.
Indeed, IL-1β induced-inflammation has been reported to be reversed by EGCG treatment in other model systems.34,35 Among the 34 RNAs we interrogated, wounding injury induced expression of 15 molecules in the IL-1β and related inflammation signaling pathway, though these failed to attain statistical significance. In the EGCG-treated group, it was found that EGCG effectively reduced the expression of 12/15 wounding-induced molecules, with the suppression of IL-1B and TNFB (or TNFα 2) attaining significance. Indeed, both IL-1 and TNFB are important mediators of inflammation.36 The expression of these two pro-inflammatory cytokines has synergistic effects, with many reports documenting IL1B inducing biological active TNFα production.37 For more than two decades, pharmacological blockage of IL-1 (ie, Anakinra, Canakinumab, etc.) constitute major treatment modalities in inflammatory diseases.38 At the same time, inhibiting TNFα has been effective in accelerating wound healing and subduing inflammation.39,40
These studies underscore the utility of the transparent Tg zebrafish as an ideal model to use for tracking migration of any tagged cells, as exemplified by the neutrophil-focused analyses presented here. Second, these studies resonate will with the accumulating literature that indicate that EGCG is potent at suppressing inflammation, by subduing IL-1, TNFB, and related pro-inflammatory molecules. Further studies are warranted to examine EGCG-induced molecular changes in this model more comprehensively, and to unravel the molecular mechanisms by which neutrophil migration is controlled. Finally, since Tg integration site can potentially affect phenotypes, it is also important to confirm these findings using additional independent Tg lines or other neutrophil tagged Tg models, in future studies.
Data Availability
All data are freely available by contacting the corresponding author.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Serhan CN, Brain SD, Buckley CD, et al. Resolution of inflammation: state of the art, definitions and terms. Faseb J. 2007;21(2):325–332. doi: 10.1096/fj.06-7227rev [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3(2):133–146. doi: 10.1038/nri1001 [DOI] [PubMed] [Google Scholar]
- 3.Weiss SJ. Tissue destruction by neutrophils. N Engl J Med. 1989;320(6):365–376. doi: 10.1056/NEJM198902093200606 [DOI] [PubMed] [Google Scholar]
- 4.Der E, Trigunaite A. Pro- and anti-inflammatory neutrophils in lupus. J Clin Cell Immunol. 2014;5(4):239. [Google Scholar]
- 5.Butterfield TA, Best TM, Merrick MA. The dual roles of neutrophils and macrophages in inflammation: a critical balance between tissue damage and repair. J Athl Train. 2006;41(4):457–465. [PMC free article] [PubMed] [Google Scholar]
- 6.Serhan CN. Novel omega – 3-derived local mediators in anti-inflammation and resolution. Pharmacol Ther. 2005;105(1):7–21. doi: 10.1016/j.pharmthera.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 7.Jin FY, Nathan C, Radzioch D, Ding A. Secretory leukocyte protease inhibitor: a macrophage product induced by and antagonistic to bacterial lipopolysaccharide. Cell. 1997;88(3):417–426. doi: 10.1016/s0092-8674(00)81880-2 [DOI] [PubMed] [Google Scholar]
- 8.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6(3):173–182. doi: 10.1038/nri1785 [DOI] [PubMed] [Google Scholar]
- 9.Pietrosimone KM, Liu P. Contributions of neutrophils to the adaptive immune response in autoimmune disease. World J Transl Med. 2015;4(3):60–68. doi: 10.5528/wjtm.v4.i3.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rossi AG, Sawatzky DA, Walker A, et al. Cyclin-dependent kinase inhibitors enhance the resolution of inflammation by promoting inflammatory cell apoptosis. Nat Med. 2006;12(9):1056–1064. doi: 10.1038/nm1468 [DOI] [PubMed] [Google Scholar]
- 11.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 12.Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (brachydanio rerio). Nature. 1981;291(5813):293–296. doi: 10.1038/291293a0 [DOI] [PubMed] [Google Scholar]
- 13.Westerfield M, Liu DW, Kimmel CB, Walker C. Pathfinding and synapse formation in a zebrafish mutant lacking functional acetylcholine receptors. Neuron. 1990;4(6):867–874. [DOI] [PubMed] [Google Scholar]
- 14.Felsenfeld AL, Walker C, Westerfield M, Kimmel C, Streisinger G. Mutations affecting skeletal muscle myofibril structure in the zebrafish. Development. 1990;108(3):443–459. [DOI] [PubMed] [Google Scholar]
- 15.Donovan A, Brownlie A, Zhou Y, et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nat. 2000;403(6771):776–781. doi: 10.1038/35001596 [DOI] [PubMed] [Google Scholar]
- 16.Montosi G, Donovan A, Totaro A, et al. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108(4):619–623. doi: 10.1172/JCI13468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. Development and maturation of the immune system in zebrafish, danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol. 2004;28(1):9–28. [DOI] [PubMed] [Google Scholar]
- 18.Lam SH, Chua HL, Gong Z, Wen Z, Lam TJ, Sin YM. Morphologic transformation of the thymus in developing zebrafish. Dev Dyn. 2002;225(1):87–94. doi: 10.1002/dvdy.10127 [DOI] [PubMed] [Google Scholar]
- 19.Bennett CM, Kanki JP, Rhodes J, et al. Myelopoiesis in the zebrafish, danio rerio. Blood. 2001;98(3):643–651. doi: 10.1182/blood.v98.3.643 [DOI] [PubMed] [Google Scholar]
- 20.Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat Immunol. 2003;4(12):1238–1246. doi: 10.1038/ni1007 [DOI] [PubMed] [Google Scholar]
- 21.Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in danio rerio. Mol Immunol. 2004;40(11):759–771. [DOI] [PubMed] [Google Scholar]
- 22.Meijer AH, Gabby Krens SF, Medina Rodriguez IA, et al. Expression analysis of the toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40(11):773–783. [DOI] [PubMed] [Google Scholar]
- 23.Pase L, Layton JE, Wittmann C, et al. Neutrophil-delivered myeloperoxidase dampens the hydrogen peroxide burst after tissue wounding in zebrafish. Curr Biol. 2012;22(19):1818–1824. doi: 10.1016/j.cub.2012.07.060 [DOI] [PubMed] [Google Scholar]
- 24.Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108(13):3976–3978. doi: 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- 25.Geraldine P, Elango R, Theodore J, Sheu J, Thomas PA. Chapter 103 - effect of epigallocatechin gallate on markers of inflammation In: Preedy VR, editor. Tea in Health and Disease Prevention. Academic Press; Waltham, MA, USA. 2013:1223–1237. [Google Scholar]
- 26.Yang F, Oz HS, Barve S, de Villiers WJ, McClain CJ, Varilek GW. The green tea polyphenol (-)-epigallocatechin-3-gallate blocks nuclear factor-kappa B activation by inhibiting I kappa B kinase activity in the intestinal epithelial cell line IEC-6. Mol Pharmacol. 2001;60(3):528–533. [PubMed] [Google Scholar]
- 27.Yamashita K, Suzuki Y, Matsui T, et al. Epigallocatechin gallate inhibits histamine release from rat basophilic leukemia (RBL-2H3) cells: role of tyrosine phosphorylation pathway. Biochem Biophys Res Commun. 2000;274(3):603–608. doi: 10.1006/bbrc.2000.3200 [DOI] [PubMed] [Google Scholar]
- 28.Donà M, Dell’Aica I, Calabrese F, et al. Neutrophil restraint by green tea: inhibition of inflammation, associated angiogenesis, and pulmonary fibrosis. J Immunol. 2003;170(8):4335–4341. doi: 10.4049/jimmunol.170.8.4335 [DOI] [PubMed] [Google Scholar]
- 29.Takano K, Nakaima K, Nitta M, Shibata F, Nakagawa H. Inhibitory effect of (−)-epigallocatechin 3-gallate, a polyphenol of green tea, on neutrophil chemotaxis in vitro and in vivo. J Agric Food Chem. 2004;52(14):4571–4576. doi: 10.1021/jf0355194 [DOI] [PubMed] [Google Scholar]
- 30.Ogryzko NV, Hoggett EE, Solaymani-Kohal S, et al. Zebrafish tissue injury causes upregulation of interleukin-1 and caspase-dependent amplification of the inflammatory response. Dis Model Mech. 2014;7(2):259–264. doi: 10.1242/dmm.013029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rider P, Carmi Y, Guttman O, et al. IL-1α and IL-1β recruit different myeloid cells and promote different stages of sterile inflammation. J Immunol. 2011;187(9):4835–4843. doi: 10.4049/jimmunol.1102048 [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi Y. The role of chemokines in neutrophil biology. Front Biosci. 2008;13:132400–132407. [DOI] [PubMed] [Google Scholar]
- 33.Guma M, Ronacher L, Liu-Bryan R, Takai S, Karin M, Corr M. Caspase-1 independent IL-1β activation in neutrophil dependent inflammation. Arthritis Rheum. 2009;60(12):3642–3650. doi: 10.1002/art.24959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, de Villiers WJS, McClain CJ, Varilek GW. Green tea polyphenols block endotoxin-induced tumor necrosis factor-production and lethality in a murine model. J Nutr. 1998;128(12):2334–2340. doi: 10.1093/jn/128.12.2334 [DOI] [PubMed] [Google Scholar]
- 35.Akhtar N, Haqqi TM. Epigallocatechin-3-gallate suppresses the global interleukin-1beta-induced inflammatory response in human chondrocytes. Arthritis Res Ther. 2011;13(3):R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kondo T, Ohshima T. The dynamics of inflammatory cytokines in the healing process of mouse skin wound: a preliminary study for possible wound age determination. Int J Legal Med. 1996;108(5):231–236. [DOI] [PubMed] [Google Scholar]
- 37.Bethea JR, Gillespie GY, Benveniste EN. Interleukin-1β induction of TNF-α gene expression: involvement of protein kinase C. J Cell Physiol. 1992;152(2):264–273. doi: 10.1002/jcp.1041520207 [DOI] [PubMed] [Google Scholar]
- 38.Dinarello CA, Simon A, van der Meer JWM. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11(8):633–652. doi: 10.1038/nrd3800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashcroft GS, Jeong M, Ashworth JJ, et al. TNFα is a therapeutic target for impaired cutaneous wound healing. Wound Repair Regen. 2012;20(1):38–49. doi: 10.1111/j.1524-475X.2011.00748.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao Y, Bao S, Yang W, et al. Epigallocatechin gallate prevents inflammation by reducing macrophage infiltration and inhibiting tumor necrosis factor-[alpha] signaling in the pancreas of rats on a high-fat diet. Nutr Res. 2014;34(12):1066. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are freely available by contacting the corresponding author.