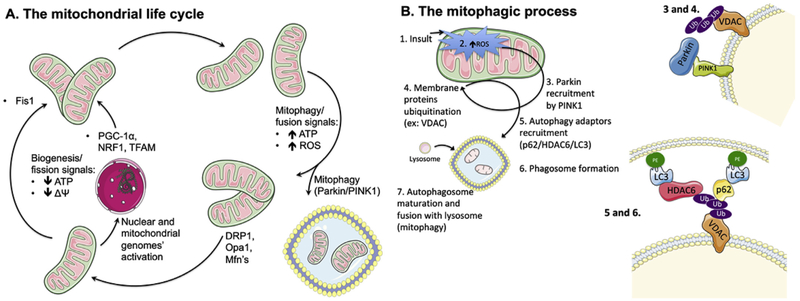
Fig. 1.
A. The mitochondrial life cycle. The mitochondrial population is a highly dynamic and fluid entity within a cell, with organelle units continually being produced or removed, depending on the cell's needs and diverse signals. One of the cell's mechanisms to cope with a decrease in cellular ATP levels, with concomitant fall in mitochondrial membrane potential (ΔΨ), is to generate more mitochondria either by fission of existing ones (which decreases membrane area per organelle, thus elevating ΔΨ) or simply producing newer ones, with resource to the genetic templates within the nucleus and the mitochondrial genome. For the biogenesis process, example of key players are PGC-1α, NRF1 and TFAM, while FIS1 is typically crucial for fission. Conversely, when ATP levels are high, oxidative stress is also elevated. As such, it becomes energetically costly to maintain numerous, unnecessary mitochondria, some of them quite damaged. As such, the cell induces either the removal of damaged mitochondria by mitophagy (which involves, for example, Parkin and PINK1, and is discussed in more detail in Fig. 1B) or fuses unnecessary mitochondria (using effectors such as DRP1, OPA1 or MFN, reducing the number of units but increasing they surface area, effectively decreasing ΔΨ and thus contributing to a lower ATP generation rate as well as decreased oxidative stress. DRP1: dynamin related protein 1, Fis1: mitochondrial fission 1 protein, Mfn: mitofusin, NRF1: nuclear respiratory factor 1, Opa1: mitochondrial dynamin-like 120 kDa protein, PINK: phosphatase and tensin homolog (PTEN)-induced kinase 1, PGC-1α: peroxisome proliferator-activated receptor gamma coactivator-1α, ROS: reactive oxygen species, TFAM: mitochondrial transcription factor A. B. Mitochondrial autophagy (mitophagy) dependency on ROS. When an insult (1) leads to elevation of mitochondrially-generated ROS levels (2), with concomitant loss of membrane potential, the membrane-bound PINK1 protein is stabilized and undergoes autophosphorylation, which triggers the recruitment of Parkin (3), a soluble E3 ubiquitin ligase. Parkin then ubiquitinates (4) other outer mitochondrial membrane proteins (such as VDAC, MFN1, and others), which is a signal for the recruitment of the autophagy adaptors p62/SQSTM1 and HDAC6 (5). These proteins anchor active LC3 units, which are themselves bound to phosphatidylethanolamine, serving as initiator point for phagosome membrane formation and maturation. Finally, the mature autophagosome is fused with hydrolytic-enzymes carrying lysosomes (7), which leads to the degradation of the lysophagosome's contents. HDAC6: histone deacetylase 6, LC3: microtubule-associated protein 1A/1B-light chain 3, MFN: mitofusin, p62/SQSTM1: p62/signaling adaptor sequestosome 1, PE: phosphatidylethanolamine, PINK1: phosphatase and tensin homolog (PTEN)-induced kinase 1, ROS: reactive oxygen species, VDAC: voltage-dependent anion channel. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)