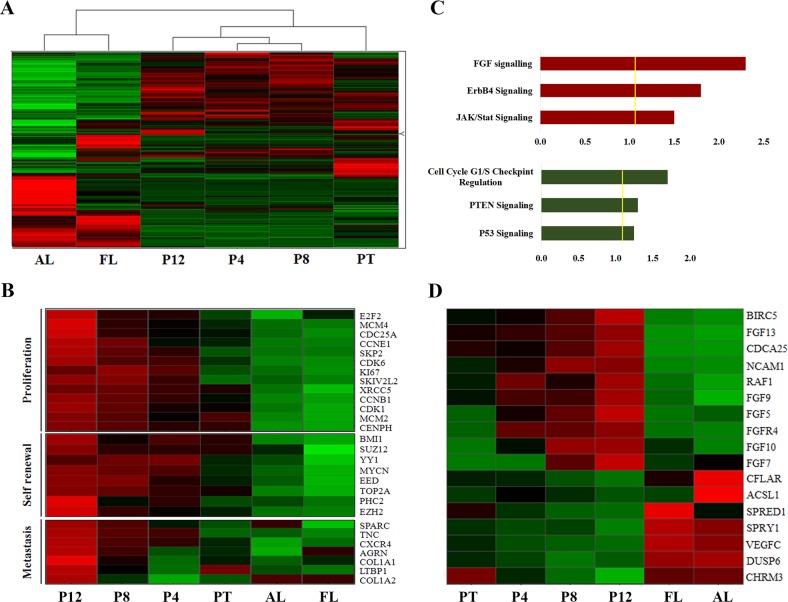
Fig. 2. Global gene signature reveals putative biomarkers involved in tumor aggressiveness.
a Microarray gene expression analysis comparing the different samples: 1. Primary PPB (PT); 2. Early PPB PDX (Passage 4-P4); 3. Intermediate PPB PDX (Passage 8-P8); 4. Late PPB PDX (Passage 12-P12); 5. Adult lung (AL); 6. Fetal lung (FL), reveals resemblance between PT and its derived Xn samples in comparison to adult and fetal normal lung tissues. b Gene heat map reveals high expression of proliferation genes in later passages (e.g., KI-67, CDK1, and E2F2), upregulation of self-renewal genes (e.g., BMI1, TOP2A, and EZH2), and an invasive gene signature (e.g., SPARC, CXCR4, and TNC) in late passage PDX. c Ingenuity® comprehensive pathway and network analysis reveals that FGF signaling pathway is one of the most upregulated pathway in late aggressive PDX passages, among the most downregulated pathways are cell cycle checkpoints regulation, PTEN and P53 pathways. d Gene heat map revealed upregulation of genes that activate the FGF signaling pathway and FGF pathway target genes (e.g., FGF5, FGF7, FGF10, BIRC5, and RAF1), as well as a low expression of genes that downregulate the FGF signaling pathway (e.g., CFLAR, SPRY1, ACSL1, and DUSP6) in late passage PDX in comparison to early passages PDX