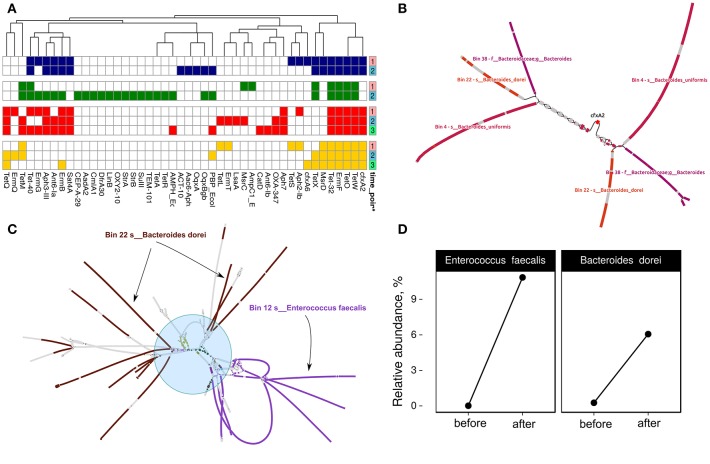
Figure 5.
Discovery of links between MAGs and antibiotic resistance genes. (A) Resistome profiling of four patients gut metagenomes. Color shows individual patients (red—HP003, blue—HP009, green—HP010, yellow—HP028). Time points are denoted by a right color bar. (B) CFX-group gene within transposon-like structure is associated with three MAGs: Bin 38—unknown Bacteroides, Bin 22—Bacteroides dorei, and Bin 4—Bacteroides uniformis. Color shows areas of the MAGs close to the graph sequences according to the BLAST results (patient HP003, time points 2 and 3). (C) Multiple sets of ARGs such as ermB, ermT, Ant6-Ia, Aph-III, Aph7, Sat4A, catA, tetL, tetM, and tetQ (shown by the blue circle and different colors) are located close to Bin 12—Enterococcus faecalis and Bin 22—B. dorei MAGs (patient HP003, time point 2). (D) Relative abundance of E. faecalis and B. dorei in HP003 patient gut metagenomes before and after the HP eradication therapy according to MetaPhlan2 analysis.