Figure 4.
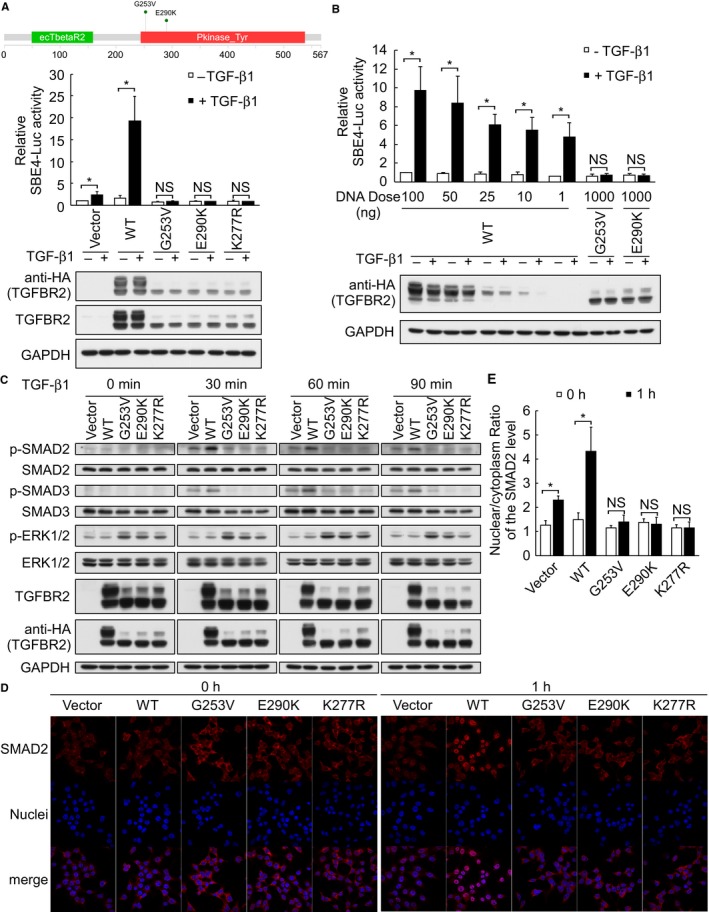
TGFBR2 G253V and E290K loss‐of‐function mutations inactivate TGF‐β/SMAD signaling. (A) Upper panel represents the schematic diagram of the TGFBR2 mutations identified in this study, as generated by the MutationMapper visualization tool. (A) and (B) HEK293T cells were cotransfected with constructs encoding HA‐tagged WT or mutant TGFBR2, SBE4‐Luc, and pRL‐TK, and replaced with serum‐free DMEM after 6 h. Cells were maintained in the serum‐free medium for another 18 h before stimulated with or without 5 ng/mL TGF‐β1 for 15 h. Promoter activities were measured using a dual luciferase assay system. Firefly luciferase activities were normalized to those of Renilla luciferase. (C) Immunoblot analysis of TGF‐β/SMAD‐ and non‐SMAD signal transduction‐related components in HEK293T cells transfected with empty vector or vectors encoding WT or mutant TGFBR2 for 24 h and then stimulated with 5 ng/mL TGF‐β1 for 0, 30, 60, or 90 min. (D) and (E) Immunofluorescence analysis of SMAD2 translocation in HEK293T cells transfected with empty vector or vectors encoding HA‐tagged WT or mutant TGFBR2 for 24 h and then stimulated with 5 ng/mL TGF‐β1 for 0 or 1 h. The nuclear/cytoplasmic ratios of SMAD2 were quantified using an IN Cell Analyzer. Values on bar graphs were shown as the mean ± SD of three independent experiments with duplicates. Intergroup comparisons were conducted by using Student's t test. (NS, not significant; *P < 0.05)
