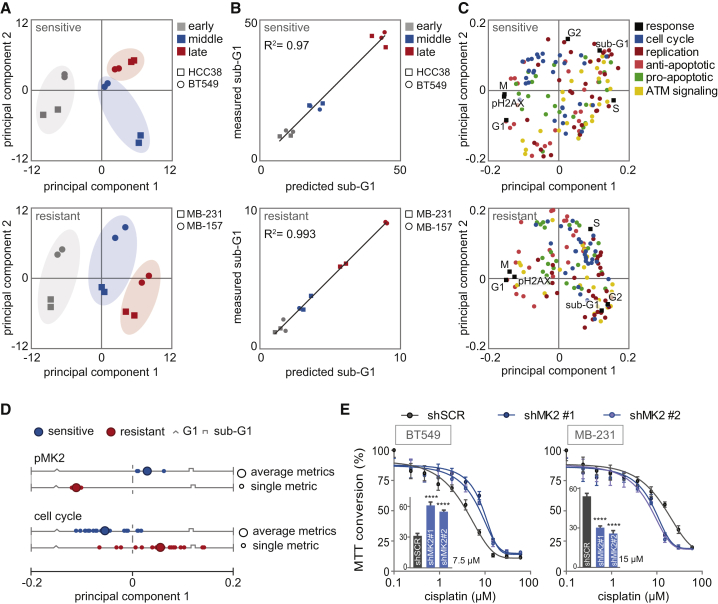
Figure 3.
PLSR Correctly Predicts Sub-G1 from Molecular Signals Activated by Cisplatin
(A) PLSR analysis of covariation between molecular signals and cellular responses. Score plots represent the signaling response of each TNBC cell line at a specified time, as indicated by the colors and symbols in the legend. Scores are plotted for the sensitive and resistant PLS models.
(B) Correlation between measured sub-G1 (flow cytometry, y axis) and model-predicted sub-G1 (x axis).
(C) PLS loadings plotted for signals and responses and colored by signaling class.
(D) PC1 loading scores of the dynamic signaling metrics (FLD, fold change; DYN, dynamic range; SMX, maximum slope; SLP, slope) are plotted. Loading scores of the four dynamic metrics of pMK2 and their average are shown in the upper panel. Loading scores of the dynamic metrics of all cell cycle-related signals (PLK1, Aurora-A, CyclinB1, CDC25C, and CDC25A) and their averages are shown in the bottom panel.
(E) Cisplatin sensitivity of BT549 and MDA-MB-231 cell lines, transduced with indicated shRNAs measured by MTT conversion. Inset bar graphs depict MTT conversion upon treatment with 7.5 or 15 μM cisplatin of BT549 and MDA-MB-231, respectively.
Error bars indicate SEM of three independent experiments. The p values were calculated using two-tailed Student’s t test. ∗∗∗∗p < 0.0001. See also Figures S3 and S4.