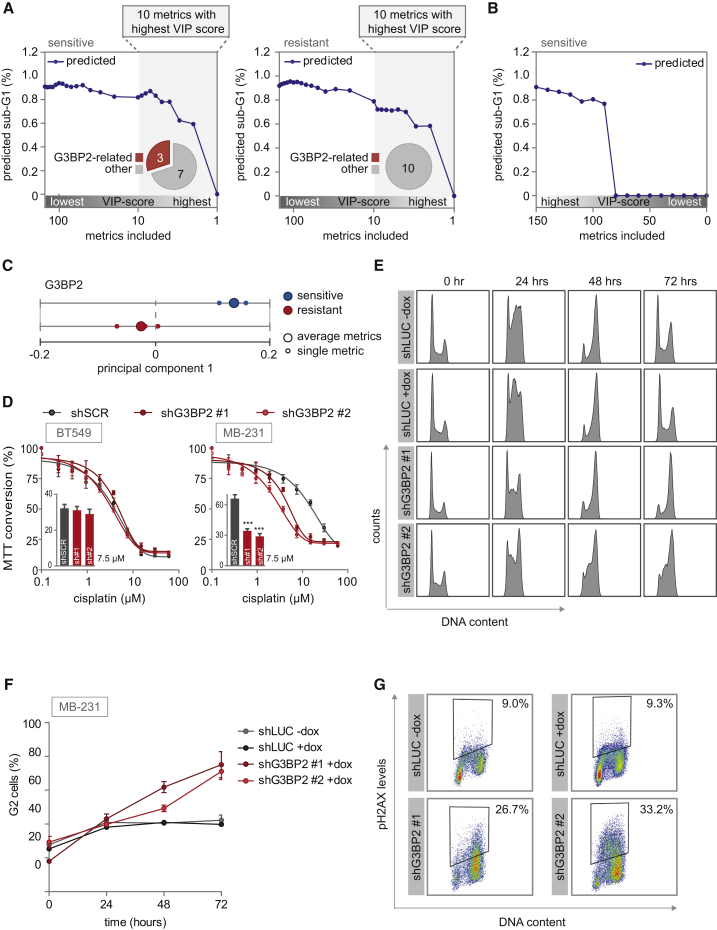
Figure 5.
Robustness of PLSR Models, and Validation of G3BP2 as a Determinant of Cisplatin Sensitivity
(A and B) The minimal number of signaling metrics required for predicting sub-G1 was calculated by iteratively removing metrics. The fraction of G3BP2-related metrics among metrics with the highest VIP scores is indicated in pie charts. (A) Metrics were eliminated sequentially from the models of cisplatin-sensitive cell lines (left panel) or cisplatin-resistant cell lines (right panel) based on the relative magnitude of their coefficients in the model, from highest to lowest VIP score. (B) Metrics were sequentially eliminated from the model of cisplatin-sensitive cell lines from lowest to highest VIP score.
(C) PC1 loading scores of the dynamic signaling metrics of G3BP2 and their average are plotted for the sensitive and resistant models individually.
(D) MDA-MB-231 and BT549 cells were transduced with indicated shRNAs and MTT conversion after cisplatin treatment was measured. Inset bar graphs depict MTT conversion upon treatment with 7.5 μM cisplatin. sh#1 and sh#2 refer to shG3BP2#1 and shG3BP2#2, respectively. Error bars indicate SEM of three independent experiments. The p values were calculated using two-tailed Student’s t test. ∗∗∗p < 0.001.
(E and F) MDA-MB-231 cells with doxycycline-inducible shRNAs targeting luciferase or G3BP2 were treated with 2 μM cisplatin. At indicated time points, cell-cycle profiles were determined by flow cytometry (E). Means and SDs of percentages of G2 cells from three independent experiments are plotted (F).
(G) γH2AX levels after 2 μM cisplatin treatment for 72 h. MDA-MB-231 cells expressing inducible shRNAs against luciferase or G3BP2 were fixed and stained with anti-γH2AX antibody and propidium iodide. γH2AX levels and DNA content were determined by flow cytometry of two independent experiments.
See also Figure S6.