Figure 3.
UCP2 Loss in AOM/DSS-Treated Mice Leads to a Differential Metabolic Gene Expression in Colon Tumors
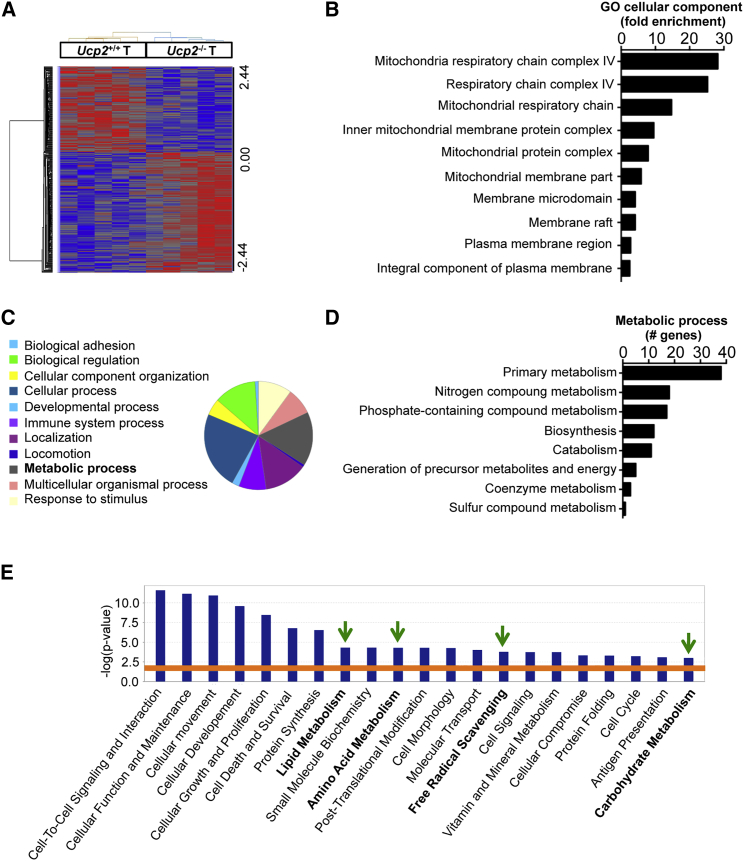
(A) Heatmap of the log2 relative expression values for each gene in each tumor (T) type plotted in red–blue color scale, with red indicating high gene expression and blue indicating low gene expression (n = 5).
(B) Bar plot ranking of the top 10 Gene Ontology (GO) cellular components based on fold enrichment.
(C) Pie chart of the biological processes associated with the upregulated genes after UCP2 loss in colon tumors. Biological categories were obtained using GO annotations from the PANTHER classification system.
(D) GO analysis of the total number of differentially expressed genes involved in metabolic processes.
(E) Gene function analysis using the software Ingenuity Pathway Analysis (IPA). Graph shows top molecular and cellular functions altered in Ucp2−/− tumors compared to Ucp2+/+ tumors. Bars above the line are statistically significant (p < 0.05) (n = 5).
See also Table S2.