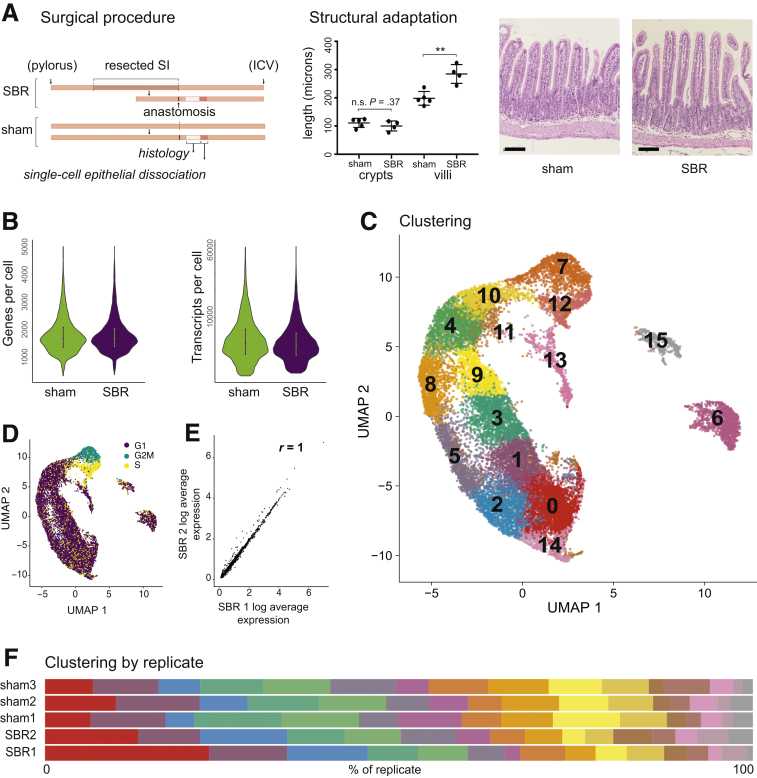
Figure 1.
Experimental design, quality control, and single-cell analysis. (A) A 50% proximal SBR and sham operation were performed on mice. Seven days after surgery, the intestine distal to the anastomosis (ileum) was harvested and equal amounts of tissue equidistant from the anastomosis were used to generate single-cell epithelial suspensions. An area immediately adjacent to this was prepared for histological examination. Typical structural adaptation of SBR mice (lengthened villi) was confirmed (P = .003), with a representative hematoxylin and eosin image of SI tissue from a sham vs SBR mouse shown (20× image acquired using Nikon Eclipse 80i). Scale bar = 100 μm. Epithelium from mice demonstrating structural adaptation was prepared for scRNA-seq analysis. (B) A mean of 1767 and 1763 genes per cell in sham and SBR, respectively, and 6754 and 6111 transcripts per cell in sham and SBR, respectively, were detected. (C) UMAP of integrated biological replicates identified 16 unique cell clusters. (D) Cell cycle states projected onto the UMAP. (E) Representative plot of SBR experimental replicates demonstrated similar gene expression profiles. Correlation coefficient (R) of average gene expression are as shown between these biological replicates. Total biological replicates were 5 sham and 4 SBR (n = 3 “sham1,” n = 1 “sham2,” n = 1 “sham3,” n = 3 “SBR1,” n = 1 “SBR2”). (F) The same 16 clusters were identified in both sham and SBR, in all replicates, as described in panel E. Distribution of all cells across clusters 0–15 (from left to right), by replicate, is shown.