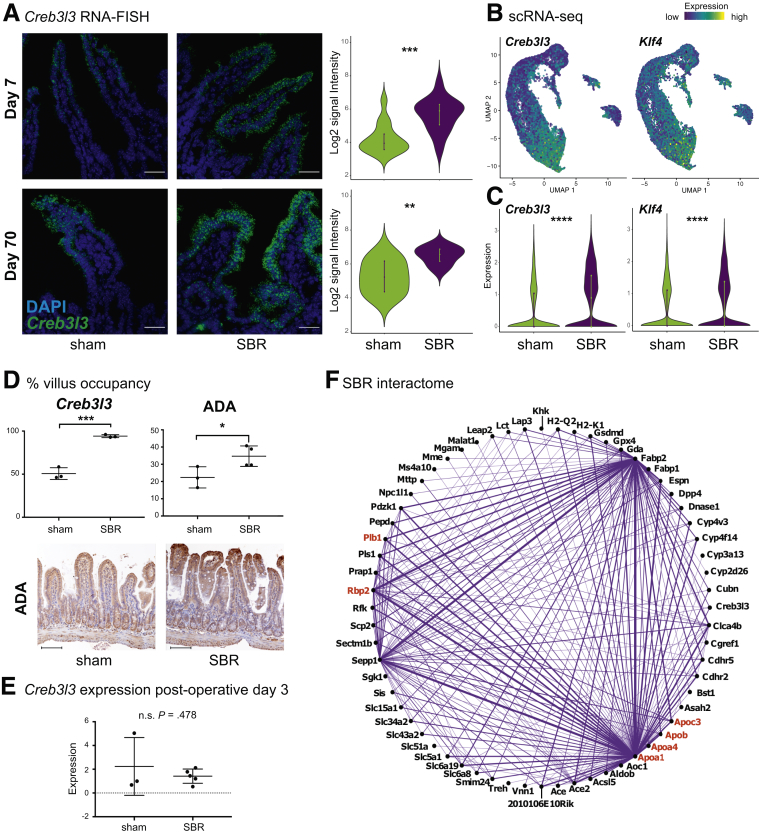
Figure 6.
Dissecting genetic underpinnings of epithelial proximalization following SBR. (A) RNA-FISH for Creb3l3 shows significant upregulation of transcripts (fluorescein signal) per nucleated cell (DAPI) at days 7 and 70 after surgery. Images are at 60×, scale bar = 30μm, acquired using Olympus FV1200 Confocal Microscope. Day 7: n = 15 sham images, n = 14 SBR images (3 biological replicates). Day 70: n = 9 sham images, n =10 SBR images (2 biological replicates). (B) Projection of Creb3l3 and Klf4 transcript enrichment onto the UMAP plot shows increased expression with enterocyte maturation. Color scale bar indicates relative intensity of gene expression. (C) Violin plots showing differential expression of Creb3l3 and Klf4 between sham and SBR. (D) The length down from the villus tip was measured for appreciably higher Creb3l3 (left, 3 sham and 3 SBR biological replicates) and ADA (right, 3 sham and 4 SBR biological replicates) expression, and represented as a percent of total villus length. Representative immunohistochemistry staining for ADA in sham and SBR mice at postoperative day 7 is shown. Images are at 20×, scale bar = 100 μm. (E) Relative Creb3l3 expression in SI from day 3 postoperative sham and SBR mice (n = 3 sham and n = 5 SBR mice) was measured using qPCR. (F) Interactome of genes upregulated in SBR epithelium. Genes in red are involved in RA signaling. All graphs are presented as mean ± SD. ns, not significant. *P < .05, **P < .01, ***P < .001, ****P < .0001.