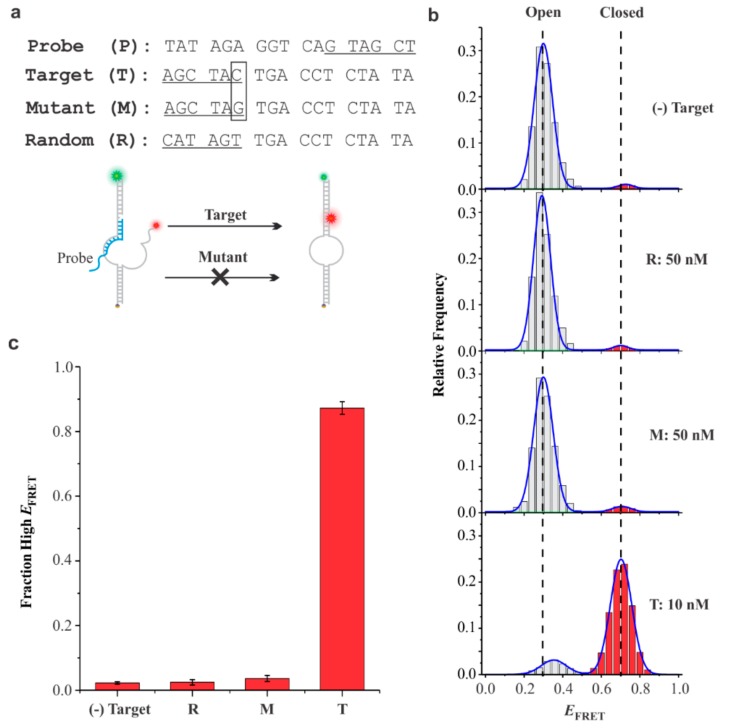
Figure 5.
Discrimination of single nucleotide mismatch from fully matched target. (a) The sequences of probe (P), target (T), mutant (M), and random (R) used for specificity analysis using sensor-III. Target refers to perfect complementary sequence. Mutant refers to a single nucleotide mutated target, whereas random refers to a target with fully randomized toehold region. The toehold regions are underlined, and the point mutation is identified with a box. (b) The smFRET histogram without the target (top panel) followed by the smFRET histograms after incubation with random, mutant, and fully matched target sequences. Almost none high-EFRET population for random and single-base mismatch target demonstrated that these sequences were not able to displace the probe. The EFRET histograms were prepared from ~200 molecules. (c) Comparison of high-EFRET fraction under different conditions. Histogram depicts that high-EFRET fractions for both the mutant and the target were similar to the background signal (2.3 ± 0.4%). However, in the presence of the target, the high-EFRET fraction increased significantly (87%). Error bars represent the standard deviations (SD), n = 3.