Kuhn et al. show that nuclear pore components Sec13 and Elys promote an open chromatin state, which can affect downstream gene expression.
Abstract
Nuclear pore complexes have emerged in recent years as chromatin-binding nuclear scaffolds, able to influence target gene expression. However, how nucleoporins (Nups) exert this control remains poorly understood. Here we show that ectopically tethering Drosophila Nups, especially Sec13, to chromatin is sufficient to induce chromatin decondensation. This decondensation is mediated through chromatin-remodeling complex PBAP, as PBAP is both robustly recruited by Sec13 and required for Sec13-induced decondensation. This phenomenon is not correlated with localization of the target locus to the nuclear periphery, but is correlated with robust recruitment of Nup Elys. Furthermore, we identified a biochemical interaction between endogenous Sec13 and Elys with PBAP, and a role for endogenous Elys in global as well as gene-specific chromatin decompaction. Together, these findings reveal a functional role and mechanism for specific nuclear pore components in promoting an open chromatin state.
Introduction
Interactions between the genome and nuclear scaffolds are known to contribute to regulation of gene expression and cell fate control, but specific mechanisms by which scaffold components influence genome regulation remain poorly defined. One of the most prominent nuclear scaffolds is the nuclear pore complex (NPC), which is known for its canonical function as a mediator of nucleocytoplasmic transport across eukaryotic nuclear membranes. In recent years, however, NPCs and their ∼30 constituent nucleoporins (Nups) have proven important for functions in genome regulation and maintenance (Raices and D’Angelo, 2017). Early EM images of mammalian nuclei have revealed decondensed chromatin preferentially associated with NPCs, interrupting the condensed heterochromatin associated with the repressive nuclear lamina. Such images have suggested a functional relationship between NPCs and open chromatin (Watson, 1955; Blobel, 1985; Capelson and Hetzer, 2009). The existence of interactions between NPCs/Nups and chromatin has now been well established in a variety of organisms via genome-wide chromatin-binding assays and imaging methods (Sood and Brickner, 2014; Ibarra and Hetzer, 2015; Ptak and Wozniak, 2016). In agreement with the EM images, the majority of these interactions were found to occur at open chromatin regions, such as actively transcribing genes (Casolari et al., 2004; Cabal et al., 2006; Capelson et al., 2010; Kalverda et al., 2010; Vaquerizas et al., 2010; Liang et al., 2013; Light et al., 2013), DNase I hypersensitive sites, and regions marked with active histone modifications such as H3K27 acetylation (Ibarra et al., 2016; Pascual-Garcia et al., 2017).
Functionally, several Nups were found to be required for the transcriptional output and regulation of at least a subset of their target genes. In metazoans, Nup targets include genes important for tissue-specific development, regulation of the cell cycle, and antiviral responses (Panda et al., 2014; Pascual-Garcia et al., 2014; Ibarra et al., 2016; Raices et al., 2017). One conserved regulatory mechanism that requires Nups is transcriptional memory, a process by which genes are marked as recently transcribed to allow more robust transcriptional responses to future activation (Light et al., 2013). Loss-of-function studies have demonstrated that specific Nups are required for multiple molecular steps involved in transcription and transcriptional memory, including binding of poised RNA polymerase (RNAP) II, H3K4 methylation, chromatin remodeling, and formation of activation-induced genomic loops (Brickner et al., 2007; Tan-Wong et al., 2009; D’Urso et al., 2016; Pascual-Garcia et al., 2017). However, while Nups have been shown to be required for these molecular events, it remains unclear which specific steps of the transcriptional or epigenetic processes Nups are sufficient to induce.
In Drosophila melanogaster, Nups such as Nup98, Sec13, and Nup62 have been detected at a large number of active genes via DNA adenine methyltransferase identification, chromatin immunoprecipitation (ChIP), and imaging studies (Capelson et al., 2010; Kalverda et al., 2010). Depletion of Sec13 or Nup98 in fly culture cells or in salivary gland tissues has been shown to lead to more compact chromatin, decreased levels of active RNAP II, and reduced mRNA production at select target genes (Capelson et al., 2010; Kalverda et al., 2010; Panda et al., 2014; Pascual-Garcia et al., 2014). Nup98 has been extensively implicated in maintaining transcriptional memory of its target genes in yeast, fly, and mammalian cells (D’Urso and Brickner, 2017), and we have recently reported that Nup98 is involved in stabilization of enhancer–promoter contacts of ecdysone-inducible genes (Pascual-Garcia et al., 2017). However, the molecular functions performed by other transcription-associated Nups such as Sec13 and Nup62 at Nup–chromatin contacts remain unknown. Additionally, many of the Nup–chromatin contacts can occur off-pore in the nuclear interior (Capelson et al., 2010; Kalverda et al., 2010; Vaquerizas et al., 2010), as these Nups have been found to shuttle NPCs on and off and/or have distinct intranuclear pools (Rabut et al., 2004). However, it remains unclear if gene regulatory functions of Nups are independent of nuclear localization.
To examine these functions and to identify which chromatin- or transcription-associated changes Nups are sufficient to induce, we used a gain-of-function approach. We generated a tethering system to create ectopic chromatin-binding sites of Sec13 and Nup62 in the genome of transgenic Drosophila strains. Using this system, we observed that NPC component Sec13 consistently induces robust chromatin decondensation at multiple genomic locations. In dissecting the mechanism of this phenomenon, we implicated Nup Elys as the primary mediator of chromatin decompaction and identified a robust interaction of Sec13 and Elys with the ATP-dependent chromatin-remodeling complex polybromo-containing Brahma (Brm)-associated proteins (PBAP), as well as a role of Elys in endogenous chromatin decondensation. These findings suggest that promoting chromatin decondensation is a critical and previously underappreciated molecular function of specific Nups in the process of gene regulation.
Results
Ectopic targeting of Nups to chromatin induces apparent chromatin decondensation at multiple genomic locations
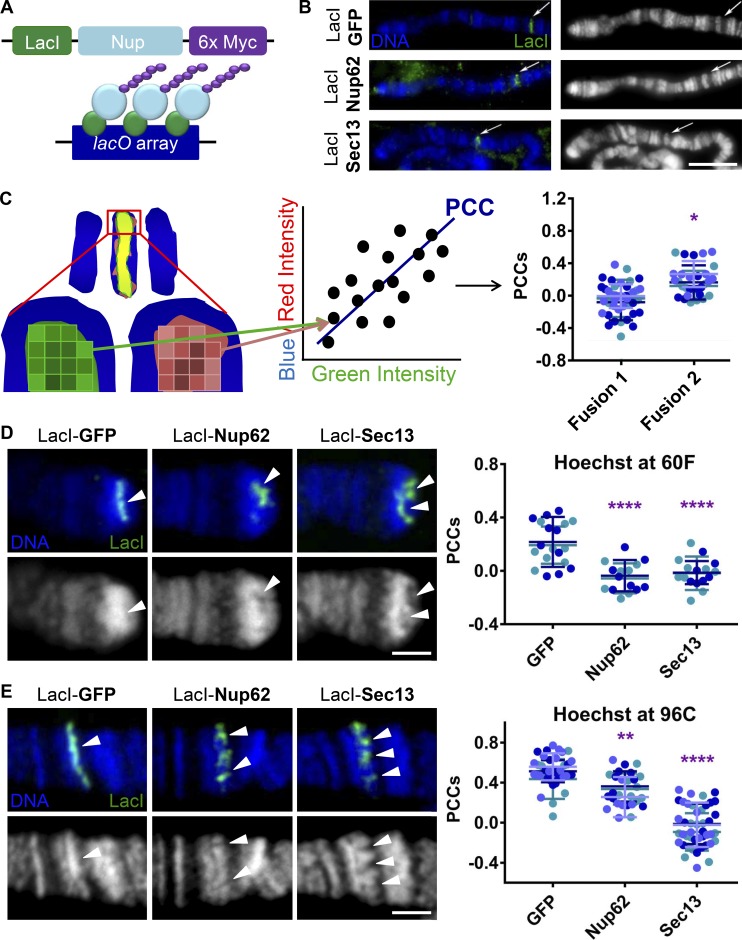
To define chromatin-related functions of Nups, and to better separate chromatin-associated roles of Nups from their transport-related functions, we used the lacO-LacI tethering system to create ectopic chromatin-binding sites of Sec13 and Nup62. We generated transgenic Drosophila melanogaster lines containing the DNA binding domain of LacI (Tumbar et al., 1999; Danzer and Wallrath, 2004) fused to either Nup62 or Sec13, under inducible control of the upstream activating sequence (UAS) element. We genetically combined these LacI-Nup lines, or a preexisting line containing a control LacI-GFP fusion (Deng et al., 2008), with a Gal4 driver expressed in third instar larval salivary glands and an integrated genomic lacO repeat array, to which the LacI-fusion proteins bind with high affinity (Fig. 1 A). We visualized this tethering using immunofluorescence (IF) of Drosophila larval salivary gland polytene chromosome squashes. These experiments allow high-resolution visualization of chromatin structure in the highly reproducible banding patterns of condensed and decondensed chromatin of the large polytene chromosomes, where we strived to identify any chromatin changes brought about by LacI-Nup fusions. We first used a lacO integration site at cytological location 4D5, which is in close proximity to the easily recognizable end of the Drosophila X chromosome, to ensure accurate detection of targeting to the lacO site. At lacO-4D5, all our LacI-fusion proteins can be reliably visualized (Fig. 1 B). Additionally, we observed correct fusion protein size by Western blotting of larval extracts (Fig. S1 A) and targeting of LacI-Nup proteins to the NPCs, as assayed by co-staining with mAb414 antibody in semi-intact salivary gland nuclei (Fig. S1 B), which is indicative of proper Nup fusion protein folding and function. Together, these data suggest a robust assay for targeting Nups to genomic loci.
Figure 1.
Ectopic targeting of Nups to chromatin induces chromatin decondensation at multiple genomic locations. (A) Schematic of lacO-LacI-Nup–inducible chromatin tethering system. (B) Widefield IF of squashed polytene chromosomes with Hoechst stain (labeled DNA and shown as blue or white/gray here and hereafter) and α-LacI (green). The right column shows Hoechst only in grayscale, whereas the left column shows the overlay of both channels. Arrows point to the lacO integration site at location 4D5 near the end of the X chromosome. LacI-fusion protein expression was driven with second chromosome Nubbin-Gal4. The scale bar is 10 µm. (C) Schematic of the PCC method of analyzing fluorescence changes, where intensities of blue Hoechst or red protein of interest and green (LacI) are measured pixel-by-pixel under green-defined LacI-band, the PCC value between blue/red and green is determined for each image, and ∼30 PCC values are measured per genotype. Example significance of P < 0.05 (*). (D) Confocal IF images of LacI-fusion proteins targeted to the subtelomeric lacO integration site on squashed polytene chromosomes at location 60F stained with Hoechst (blue or white) and α-LacI (green). The top row shows the overlay of both channels, and the bottom row shows Hoechst only. “Holes” (areas of highly reduced staining density) in Hoechst staining can be reproducibly observed under LacI-Nup binding. Expression was driven with Sgs3-Gal4. Arrowheads show observed decondensation or lack thereof under LacI. The scale bar is 2 µm. Quantification displays PCCs between blue and green signal under LacI. Data are from two biological replicates (colored), each from an independent experiment. GFP, n = 19; Nup62, n = 15; Sec13, n = 17. ****, P < 0.0001. Error bars represent SDs. (E) Experimental conditions, staining, and imaging are identical to D above, with the replacement of cytological location 60F with location 96C and Nubbin-Gal4 driver. Holes in Hoechst can reproducibly be observed under LacI-Sec13 and occasionally under LacI-Nup-62. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 39; Nup62, n = 27; Sec13, n = 44. ****, P < 0.0001; **, P < 0.01. Error bars represent SDs. The image for control LacI-GFP is the same as the image for control LacI-GFP in Fig. 2 A.
Since we aimed to assay for chromatin changes associated with active transcription, we turned away from the lacO-4D5 integration site as it corresponded to an already highly decondensed and transcribing genomic locus (Fig. 1 B and data not shown). Instead, we next used a lacO integration site at cytological location 60F, a subtelomeric locus found in a highly condensed region of chromatin at the end of chromosome 2R. To interrogate changes in chromatin structure or protein recruitment in an unbiased and accurate way, we devised a highly sensitive and semi-automated method by which the fluorescent signals at the lacO site were analyzed (Fig. 1 C). The intensity of green fluorescence signal (LacI) was compared with the intensity of blue fluorescence signal (Hoechst DNA stain) or red fluorescence signal (proteins of interest) on a pixel-by-pixel basis for the area under the LacI-defined band. The Pearson correlation coefficient (PCC) representing the overall relationship of green to blue/red intensity was then calculated for each lacO site per image. By obtaining PCC measurements from many cells per gland from multiple animals, we can effectively compare differences in chromatin density or recruitment of proteins of interest between LacI-GFP control, LacI-Nup62, and LacI-Sec13 bound to lacO loci (Fig. 1 C).
Using this method, we observed a positive correlation between Hoechst signal and LacI-GFP, representing the bright DNA staining and highly condensed nature of chromatin at the subtelomeric lacO-60F site under control conditions (Fig. 1 D). However, we could visualize a striking loss of DNA signal intensity associated with binding of LacI-Nup62 or LacI-Sec13, represented by a quantifiable and significant reduction in the PCCs between LacI fusion protein and Hoechst DNA stain (Fig. 1 D). This decrease in the correlation between bound LacI-Nups and DNA signal intensity at lacO-60F suggests that chromatin becomes less dense upon LacI-Nup targeting, and implies that tethering nuclear pore proteins Nup62 or Sec13 to a genomic site is sufficient to induce chromatin decondensation.
To corroborate that the changes we observe in DNA signal intensity are associated with chromatin decondensation at this subtelomeric integration site, we stained for Drosophila telomere capping protein HP1/ORC-associated protein (HOAP), which is known to bind heterochromatin at chromosome ends (Cenci et al., 2003). We found that targeting LacI-Nup62 or LacI-Sec13 to the lacO-60F locus results in a dramatically reduced area of HOAP signal at the 2R telomere compared with control (Fig. S1 C). These images also illustrate that, in some instances, the apparent decondensation by Sec13 can be so severe that the entire telomeric end of the chromosome appears to have been decondensed, revealed by the LacI-Sec13 signal appearing at the distal-most tip of the visible DNA signal compared with the more proximal location of the band of LacI-GFP (Fig. S1 C). These data support the notion that there is a loss of the condensed heterochromatic state at the lacO-60F site upon Nup62 or Sec13 tethering.
To determine if this phenotype is reproducible, we next tethered the LacI-fusion proteins to a lacO integration site at cytological location 96C, which is a nontelomeric condensed band on chromosome 3R. Here we again observed significant loss in DNA stain fluorescence signal density associated with binding of LacI-Nup62 or LacI-Sec13 compared with LacI-GFP control, and a corresponding significant reduction in PCC values (Fig. 1 E). Interestingly, Sec13 induces the apparent chromatin decondensation much more robustly than Nup62 at the lacO-96C locus. The difference in the magnitude of observed change in chromatin structure between Sec13 and Nup62 at lacO-96C provided an opportunity to further probe the mechanism of this Nup-induced phenomenon in later experiments, as it allowed for assessing a dose-dependent relationship.
Nup binding to chromatin is associated with a decrease in histone density and an increase in gene expression
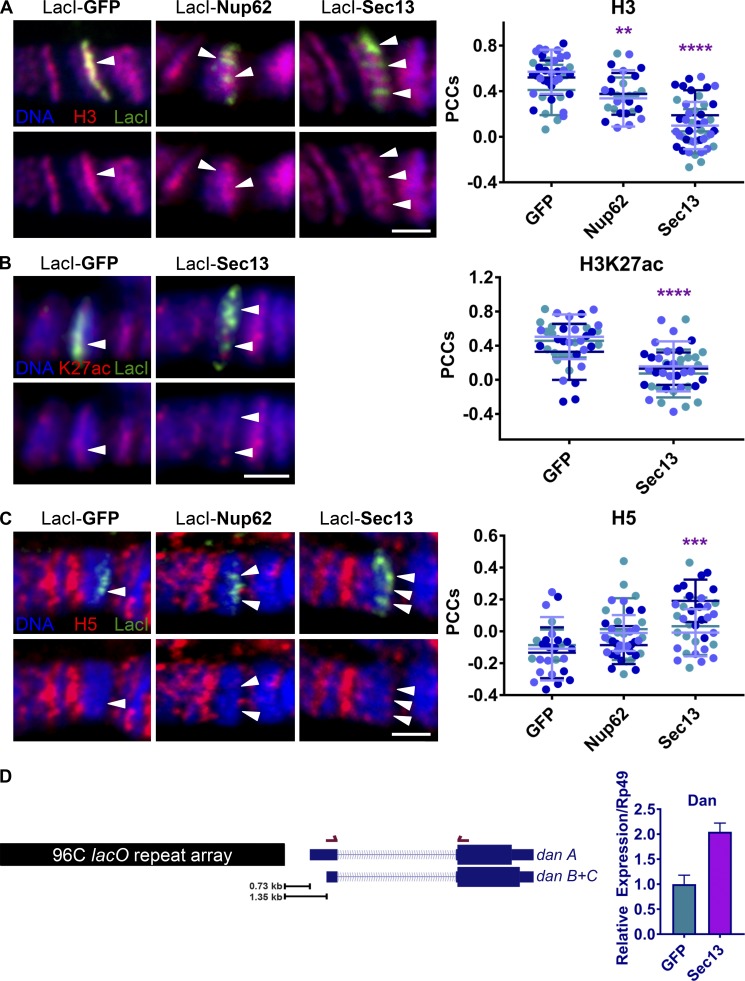
The loss of Hoechst intensity at the lacO sites upon Nup tethering suggested that chromatin is becoming less dense or more decondensed. This change in DNA stain intensity can come from the loss of nucleosomal density and/or be associated with specific histone modifications linked to active chromatin. To examine these possibilities and to further validate our conclusion that Nup tethering induces chromatin decondensation, we stained for the core histone H3 and observed a significant decrease upon Nup62 and, more robustly, Sec13 binding (Fig. 2 A). The observed decrease in histone density upon Nup tethering supports the notion that the loss of Hoechst staining, reported above (Fig. 1), represents remodeling or loss of nucleosomes. Furthermore, the difference in magnitude of H3 staining loss between Nup62 and Sec13 corresponds well with the difference in the observed Hoechst staining loss at lacO-96C between the Nups (Fig. 1 E). Next, we determined if accumulation of histone modifications associated with active transcription, such as H3K27 acetylation or H3K4 dimethylation, correlated with Sec13-induced chromatin decondensation. Interestingly, we did not observe an increase in the association of either active mark with LacI-Sec13 relative to LacI-GFP control, and instead detected a significant decrease in visible levels of both histone modifications upon Sec13 tethering (Figs. 2 B and S2 A), which is consistent with a reduction in general nucleosome occupancy at lacO-96C upon Sec13 binding (Fig. 2 A).
Figure 2.
Nup binding to chromatin is associated with a decrease in histone density and an increase in gene expression. (A) Confocal IF images of LacI-fusion proteins targeted to the lacO integration site on squashed polytene chromosomes at location 96C. Staining was with Hoechst (blue) and antibodies against H3 (red) and LacI (green). LacI-fusion protein expression was driven with Nubbin-Gal4. The top row shows the overlay of all three colors, whereas the bottom row shows blue and red only (here and in B and C). Arrowheads indicate locations of existing or depleted H3 under LacI signal. The scale bar is 2 µm. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 39; Nup62, n = 27; Sec13, n = 44. ****, P < 0.0001; **, P < 0.01. Error bars represent SDs. The image for control LacI-GFP is the same as the image for control LacI-GFP in Fig. 1 E, demonstrating enrichment of histone H3 and high Hoechst staining density at the same control locus. (B) Experimental conditions and strains are as in A above, but with H3K27ac antibody (red) instead of H3 and GFP or myc antibodies (green) instead of LacI due to antibody animal source constraints, and with the use of widefield microscopy. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 38; Sec13, n = 40. ****, P < 0.0001. Error bars represent SDs. (C) Experimental conditions and strains as in A above, but with antibodies against LacI (green) and CTD tail Ser2 phosphorylated RNAP II (H5, red), and with the use of widefield microscopy. Arrowheads indicate LacI signal and recruitment or lack thereof of H5. The scale bar is 2 µm. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 26; Nup62, n = 40; Sec13, n = 35. ***, P < 0.001. Error bars represent SDs. (D) Schematic of the distance between integration of the lacO repeat plasmid and the downstream isoforms of the dan gene along with location of the primer set used for RT-qPCR. Three technical replicates of each of three biological replicates (10 sets of glands per replicate) were used for quantification. Error bars represent SEMs.
Chromatin decondensation is a critical step in facilitating transcription factor and RNAP II binding, as well as in subsequent steps of gene transcription. RNAi-mediated depletion of Sec13 in these cells has been previously shown to result in a loss of chromatin decondensation, along with concurrent reduction of RNAP II levels and of gene expression at endogenous Sec13 targets (Capelson et al., 2010). Thus, we next wanted to determine whether Nup-induced decondensation at the ectopic site resulted in any transcription-associated changes as well.
To determine if RNAP II is recruited to the decondensed lacO-96C locus upon Nup tethering, we stained with the H5 antibody, which recognizes the serine 2 phosphorylated form and represents actively transcribing RNAP II (Phatnani and Greenleaf, 2006). We observed a modest but significant accumulation of the Ser2Ph form of RNAP II at lacO-96C when bound by Sec13 (Fig. 2 C). We next conducted quantitative RT-PCR (RT-qPCR) to measure expression levels of the dan gene, which is located ∼1.3 kb downstream of the lacO-96C integration site (Fig. 2 D and Wallrath, L.L., personal communication). We found a twofold increase in dan expression specifically when LacI-Sec13 was targeted to lacO-96C, relative to LacI-GFP control (Fig. 2 D). Together, these results suggest that chromatin decondensation associated with binding of Sec13 at this locus allows for a small but significant amount of transcriptional machinery to bind and productively transcribe downstream genes.
Nup-induced decondensation of chromatin is not associated with a change in nuclear localization
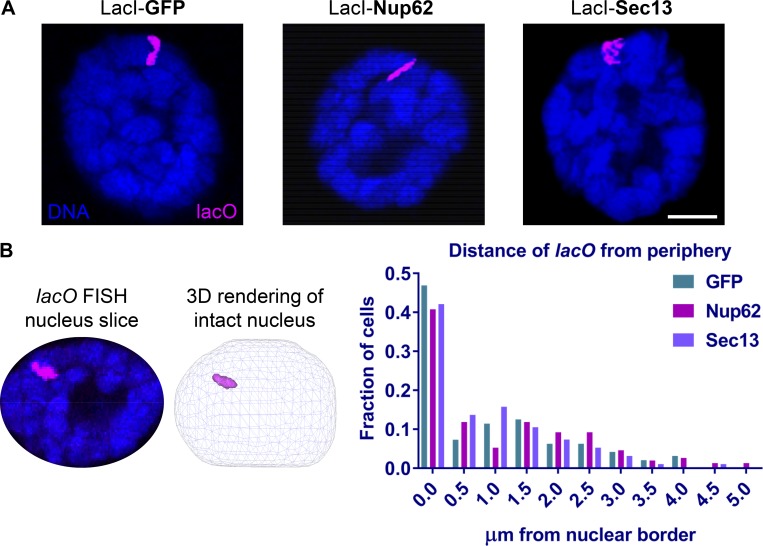
Metazoan Nups have been found to interact with chromatin both at and away from NPCs (Capelson et al., 2010; Kalverda et al., 2010; Vaquerizas et al., 2010), and many Nups demonstrate short residence times at NPCs, suggesting dynamic behaviors (Rabut et al., 2004). In light of this, we aimed to determine if ectopically chromatin-tethered Nups target the lacO-96C locus to NPCs at the nuclear periphery and if NPC association is correlated with chromatin decondensation.
To assess this, we conducted DNA FISH with fluorescently tagged oligonucleotide probes complementary to the lacO-96C locus in intact nuclei of salivary glands in our system, followed by 3D analysis of the nuclear position of the lacO probe relative to the nuclear periphery (Fig. 3, A and B). Although the lacO locus in all genotypes showed peripheral localization bias, we observed no significant difference in the percentage of peripheral (<0.5 µm from the nuclear border) lacO loci when bound by LacI-GFP, LacI-Nup62, or LacI-Sec13 (Fig. 3 B). Since Sec13 induces robust decondensation of chromatin at lacO-96C while the level of decondensation achieved by tethering Nup62 is significantly less (Fig. 1, D and E), the lack of difference in peripheral localization between either of these or the GFP control suggests that the ability of chromatin-bound Nups to induce decondensation is independent of nuclear positioning. Although polytene chromosomes are reported to be relatively immobile (Hochstrasser and Sedat, 1987), we conclude from our data that recruitment to the nuclear periphery does not appear to correlate with chromatin decondensation.
Figure 3.
Nup-induced decondensation of chromatin is independent from localization to the nuclear periphery. (A) Representative images of DNA FISH (magenta) against the lacO array at 96C in intact salivary gland polytene nuclei, stained with Hoechst (blue), obtained using 3D confocal microscopy. The scale bar is 10 µm. (B) The TANGO plugin (Ollion et al., 2013) in FIJI image analysis software (Schindelin et al., 2012) was used to compile 3D renderings of confocal Z-stacks of nuclei, call nuclear and lacO objects, and calculate minimum 3D distances of the edge of the lacO locus “object” to the edge of the Hoechst DNA-defined nuclear periphery when bound by different LacI-fusion proteins. Distances of lacO to periphery were plotted to show the fraction of cells in the salivary glands of three biological replicates (>80 cells total) per genotype from two independent experiments with distance bins in increments of 0.5 µm.
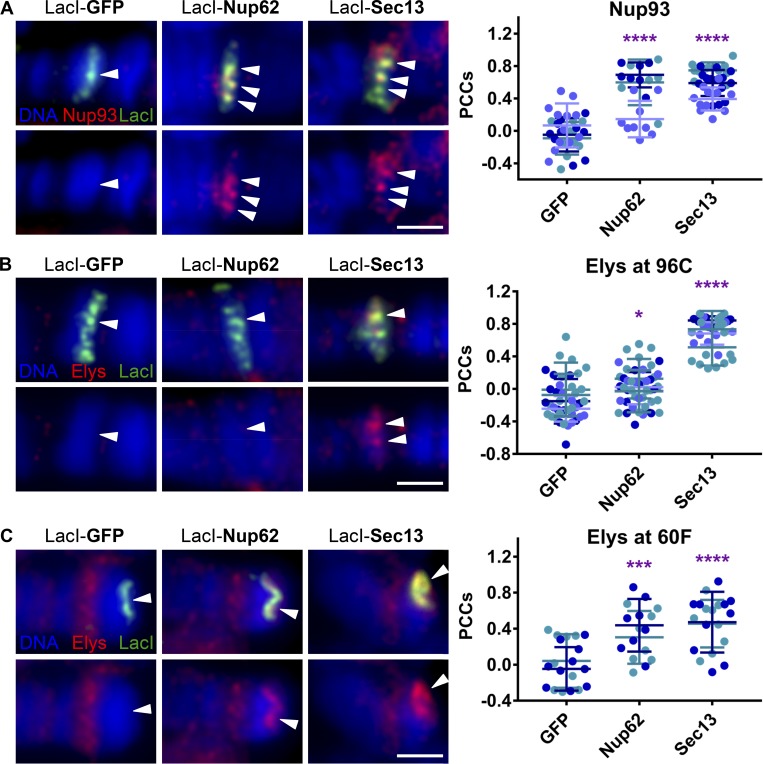
Nup-induced chromatin decondensation correlates with recruitment of Nup Elys
To further characterize Nup-induced chromatin decondensation, we aimed to determine what other NPC components are recruited by chromatin-tethered Nup62 or Sec13. We observed that both Nup62 and Sec13 recruit stable core NPC component Nup93 to lacO-96C at comparable levels (Fig. 4 A), further suggesting similar levels of interaction with peripheral NPCs (Fig. 3). However, we did observe differential and highly robust recruitment of another Nup, Elys, by Sec13 at lacO-96C (Fig. 4 B). Elys is the only Nup with a clearly defined chromatin-binding domain and activity (Zierhut et al., 2014). Our highly sensitive PCC quantification method also detected a mild recruitment of Elys by LacI-Nup62 at lacO-96C; however, this is dramatically less than the amount recruited by LacI-Sec13. Furthermore, we did not observe either Nup62 or Sec13 recruiting core NPC component Nup107 (against which we have recently generated an antibody; Fig. S2, B and C) or nuclear basket Nup Mtor to lacO-96C (Fig. S2 D), supporting the specificity of the relationship between Elys and Sec13 at lacO-96C.
Figure 4.
Nup-induced chromatin decondensation correlates with recruitment of Nup Elys. (A) Widefield IF images of LacI-fusion proteins targeted to the lacO integration site on squashed polytene chromosomes at location 96C. Staining was with Hoechst (blue) and antibodies against Nup93 (red) and LacI (green). The top row shows the overlay of all three colors, whereas the bottom row shows blue and red only (here and in B and C). Arrowheads indicate locations of observed Nup93 recruitment or lack thereof under LacI signal. The scale bar is 2 µm. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 32; Nup62, n = 22; Sec13, n = 37. ****, P < 0.0001. Error bars represent SDs. (B) Experimental conditions, strains, and imaging are as in A above, but with antibodies against Elys (red) and myc (green). Arrowheads indicate locations of observed Elys recruitment or lack thereof under LacI signal. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 42; Nup62, n = 45; Sec13, n = 40. ****, P < 0.0001; *, P < 0.05. Error bars represent SDs. (C) Experimental conditions and imaging are as in A above, but with antibodies against Elys (red) and myc (green), and at location 60F with Sgs3-Gal4 driver. Data are from two biological replicates (colored) from two independent experiments. GFP, n = 20; Nup62, n = 16; Sec13, n = 19. ****, P < 0.0001; ***, P < 0.001. Error bars represent SDs.
Given the observed correlation between recruitment of Elys and decondensation, we further probed whether the amount of Elys recruited to chromatin correlates with the degree of Nup-induced decondensation at another locus. To do so, we assessed Elys recruitment to the subtelomeric lacO-60F locus, where Nup62 induces chromatin decondensation to a level more comparable to that of Sec13 (Fig. 1 D). Strikingly, both Nup62 and Sec13 recruit significantly high and, importantly, more comparable levels of Elys to this locus, where they both decondense robustly (Fig. 4 C). These results demonstrate that the amount of decondensation in these assays correlates strongly with levels of Elys recruitment, and suggest a possible causal relationship between the two.
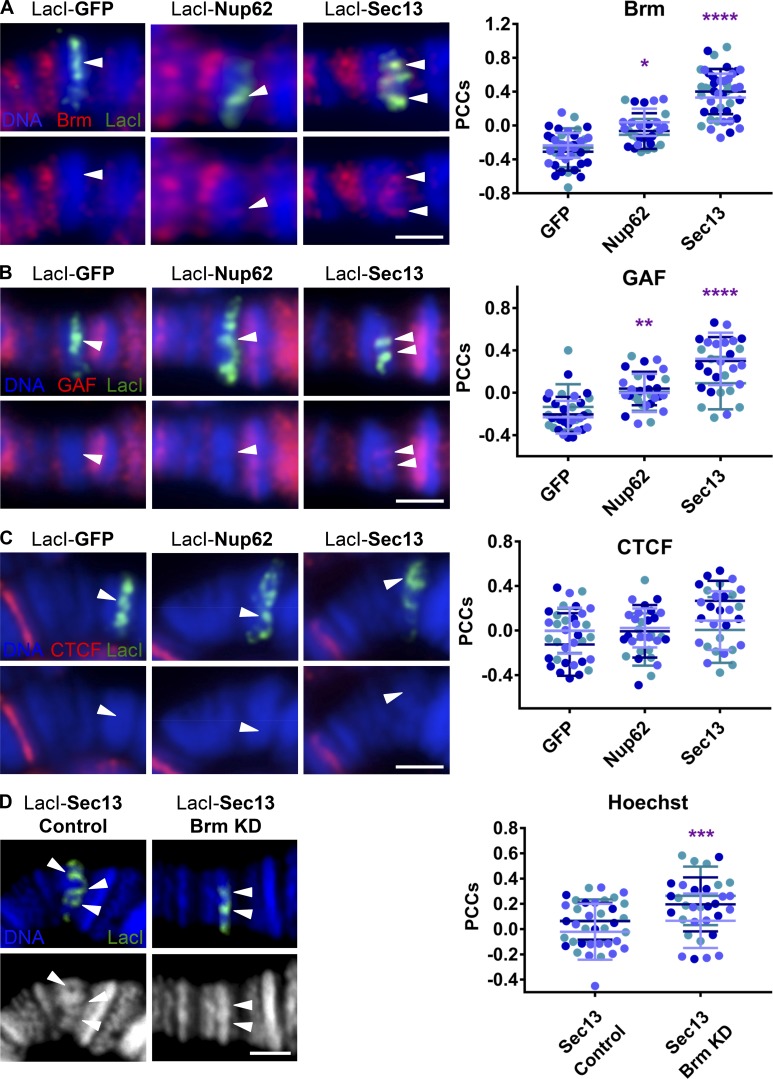
Chromatin-tethered Sec13 recruits the chromatin-remodeling PBAP/Brm complex and associated GAGA factor
To understand the molecular mechanism behind Sec13-induced decondensation, we next turned to chromatin-remodeling complexes, as they are the known enzymatic drivers of chromatin decompaction (Tyagi et al., 2016). PBAP is a Drosophila ATP-dependent switch/sucrose nonfermenting (SWI/SNF) chromatin-remodeling complex comprising nine proteins, including Brm, the ATPase, and polybromo, the specific protein that distinguishes PBAP from the related Brm-associated protein (BAP) complex (Mohrmann and Verrijzer, 2005). Strikingly, both of these proteins were significantly recruited by Sec13 to lacO-96C, most robustly Brm (Fig. 5 A and Fig. S3 A). As with Elys, a small increase in correlation between Brm and Nup62 is detected by the PCC method, but again this is significantly less than that recruited by Sec13 and closer to the levels of control GFP fusion protein. This lower level of recruitment correlates with the lower level of Nup62-induced decondensation at this locus, suggesting a dose-dependent relationship between Brm and chromatin decondensation (Fig. 1 D). These results suggest that the Nup-induced chromatin decondensation at lacO-96C is facilitated by the chromatin-remodeling complex PBAP.
Figure 5.
Chromatin-tethered Sec13 recruits the chromatin-remodeling PBAP/Brm complex and associated GAGA factor. (A) Widefield IF images of LacI-fusion proteins targeted to the lacO integration site on squashed polytene chromosomes at cytological location 96C. Staining was with Hoechst (blue) and antibodies against Brm (red) and GFP (green) for control or myc (green) for Nup fusion constructs due to antibody animal source constraints. The top row shows the overlay of all three colors, whereas the bottom row shows blue and red only (here and in B and C). Arrowheads indicate locations of observed Brm recruitment or lack thereof under LacI signal. The scale bar is 2 µm. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 41; Nup62, n = 30; Sec13, n = 45. ****, P < 0.0001; *, P < 0.05. Error bars represent SDs. (B) Experimental conditions, strains, and imaging are as in A above, but with antibodies against GAF (red) and GFP (green) for control or myc (green) for Nup fusion constructs due to antibody animal source constraints. Arrowheads indicate locations of observed GAF recruitment or lack thereof under LacI signal. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 32; Nup62, n = 29; Sec13, n = 28. ****, P < 0.0001; **, P < 0.01. Error bars represent SDs. (C) Experimental conditions, strains, and imaging are as in A above, but with antibodies against CTCF (red) and GFP (green) for control or myc (green) for Nup fusion constructs due to antibody animal source constraints. Arrowheads indicate LacI signal. The scale bar is 2 µm. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 34; Nup62, n = 30; Sec13, n = 29. Error bars represent SDs. (D) Confocal IF images of LacI-Sec13 targeted to the lacO integration site on squashed polytene chromosomes at location 96C under control conditions (flies crossed to w1118 WT stock) or Brm KD conditions (flies crossed to Brm RNAi stock BL35211). Staining was with Hoechst (blue or white) and α-LacI (green). LacI-Sec13 protein expression and Brm RNAi were driven with Nubbin-Gal4. The top row shows the overlay of the two channels, whereas the bottom row shows DNA stain only in white/grayscale. Arrowheads indicate locations of observed decondensation or lack thereof under LacI signal. The scale bar is 2 µm. Quantification displays PCCs between red and blue signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 27; Sec13, n = 33. ***, P < 0.001. Error bars represent SDs.
Interestingly, one protein previously shown to interact with PBAP, GAF (Nakayama et al., 2012), was recently found to associate with Nups in Drosophila cells (Pascual-Garcia et al., 2017). GAF is known to both play an architectural genome-organizing role and to regulate formation of DNase hypersensitive sites (Ohtsuki and Levine, 1998; Fuda et al., 2015). Thus, we assessed recruitment of GAF in our system and found GAF to be significantly recruited by Sec13 to lacO-96C, compared with control GFP or Nup62 (Fig. 5 B). To further verify specificity of proteins recruited by Sec13 to lacO-96C, we stained for architectural protein CTCF, which was also previously found to associate with Nups in certain conditions (Pascual-Garcia et al., 2017). Strikingly, the absence of CTCF at the lacO-96C under control conditions is maintained under conditions of Nup62 or Sec13 targeting (Fig. 5 C), supporting specificity of GAF and Brm recruitment by Sec13.
To investigate whether Sec13-induced chromatin decondensation indeed requires the PBAP complex, we introduced a Brm RNAi construct into our genetic tethering system. As validation, we observed that levels of Brm recruited to lacO-96C by tethered Sec13 were in fact reduced in the presence of Brm RNAi (Fig. S3 B). Analysis of Hoechst fluorescence levels at this locus yielded a visible and measureable increase in the correlation between LacI-fusion protein and Hoechst intensity levels in the presence of Brm RNAi, indicative of increased DNA density and reduced chromatin decondensation (Fig. 5 D). This result provides strong evidence that the observed robust recruitment of Brm, the ATPase component of the PBAP chromatin-remodeling complex, is responsible for the Nup-induced chromatin decondensation.
Endogenous Elys associates with Drosophila PBAP and regulates chromatin compaction
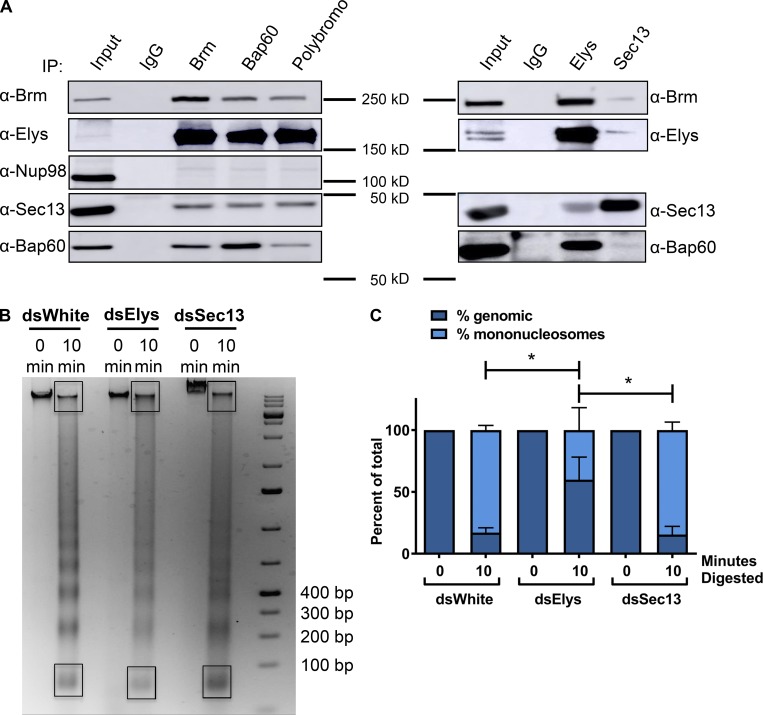
To confirm that the relationship between Nups and chromatin-remodeling proteins in our ectopic system is representative of their endogenous interactions, we conducted coimmunoprecipitation experiments in Drosophila S2 embryonic cultured cells. Immunoprecipitation of endogenous Sec13 and Elys, using previously characterized antibodies (Pascual-Garcia et al., 2017), resulted in a robust pull-down of PBAP components Brm and Bap60, especially in the case of Elys (Fig. 6 A). The reverse coimmunoprecipitation of PBAP components Brm, Bap60, and polybromo demonstrated a reciprocal interaction with Sec13 and, again even more strongly, with Elys (Fig. 6 A). Interestingly, components of PBAP did not pull down Nup98, showing specificity of this interaction. These data indicate that endogenous Sec13 and Elys physically associate with PBAP chromatin-remodeling proteins, and, based on the strength of these interactions, that Elys may be the primary interacting partner of chromatin remodelers. This conclusion is supported by our observation that there is a strong correlation between the level of Elys recruited by Nup62 (Fig. 4, B and C) and the degree to which Nup62 tethering decondenses chromatin at the two lacO loci, 96C and 60F (Fig. 1, D and E). Additionally, this is supported by the similarity between levels of recruitment of Elys and Brm by Nups at lacO-96C (Figs. 4 B and 5 A). These results support a dose-dependent relationship, where levels of recruitment of Elys, and consequently, levels of Brm, regulate the degree of Nup-induced chromatin decondensation. Together, they point to Elys as the primary mediator of chromatin decondensation driven by Nups.
Figure 6.
Endogenous Elys associates with Drosophila PBAP and regulates chromatin compaction. (A) Right: Coimmunoprecipitation experiments in S2 cell lysates, in which immunoprecipitates of the components of the PBAP complex were Western blotted for Elys, Sec13, and Nup98. Left: Coimmunoprecipitation experiments, in which immunoprecipitates of Elys or Sec13 were Western blotted for components of the PBAP complex. 10% of lysate relative to immunoprecipitate was loaded for inputs, 40% per sample. The experiment was done three times, and representative blots are shown. (B) Representative gel image of genomic DNA subjected to MNase digestion for indicated lengths of time from S2 cells treated with dsWhite (control), dsSec13, or dsElys RNAi (for 6 d). Black boxes indicate the mononucleosome band, used in quantification of digestion (in C), relative to the undigested genomic band at the top. (C) Quantification of Mnase digestion of chromatin harvested from S2 cells treated with control, Elys, or Sec13 dsRNA, displayed as a plot of relative amounts of the detected mononucleosome band and the undigested genomic band, at the indicated times of digestion. The mean and standard error bars are calculated from four independent biological replicates (two replicates from two independent experiments). *, P < 0.05.
To further explore this, we wanted to determine whether Nups also contribute to chromatin decompaction in an endogenous context. Therefore, we tested whether Sec13 and/or Elys are required for proper global nucleosome compaction, as assayed by genomic accessibility to micrococcal nuclease (MNase) digestion, in Drosophila S2 cells. RNAi-mediated reduction of Elys versus control (Fig. S4 A), using double-stranded RNA (dsRNA), resulted in a reproducibly lower ratio of mononucleosomes to undigested genomic DNA upon MNase treatment (Fig. 6, B and C), indicative of more condensed chromatin upon Elys depletion. Interestingly, RNAi depletion of Sec13 did not manifest the same phenotype (Fig. 6, B and C), suggesting that Elys is the primary facilitator of chromatin decondensation. This is consistent with the stronger interaction of Elys with PBAP components compared with Sec13 in these cells (Fig. 6 A) and is also in agreement with our previously published ChIP sequencing (ChIP-seq) profile showing binding of Elys to thousands of actively marked loci in fly tissues (Pascual-Garcia et al., 2017), further supporting the notion that Elys promotes chromatin accessibility throughout the genome.
Elys regulates levels of chromatin compaction and gene expression at endogenous gene targets
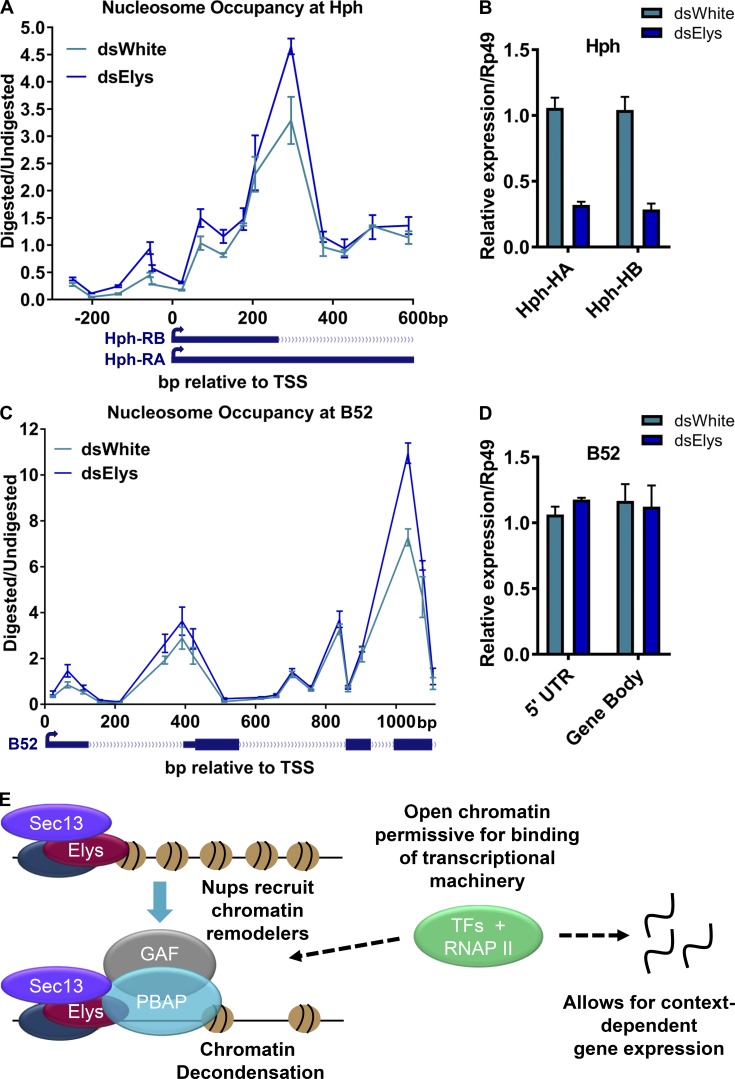
To further characterize the regulation of chromatin compaction by Elys, we analyzed its proposed functions at endogenous target genes in S2 cells. Nups have been previously shown to bind and regulate expression of Drosophila genes Hph (Pascual-Garcia et al., 2014) and B52 (Panda et al., 2014) in these cells, where both of these genes are expressed. Additionally, we have detected robust binding peaks of Elys at these genes in previous ChIP-seq experiments in fly tissues, where the Elys antibody has been shown to be specific and amenable for ChIP use (Pascual-Garcia et al., 2017). We confirmed robust binding of Elys to Hph and B52 promoter regions, relative to a negative control region (selected on the basis of lack of Elys ChIP-seq signal in fly tissues [Pascual-Garcia et al., 2017]), by quantitative ChIP-PCR (ChIP-qPCR; Fig. S4, B and C). To determine whether Elys exerts an effect on chromatin compaction of Hph and B52, we used an MNase digestion followed by quantitative PCR (qPCR; MNase-qPCR) to define occupancy levels of nucleosomes at specific loci. To verify our MNase-qPCR assay, we first tested it on a well-studied Drosophila hsp70 gene that becomes highly activated and decondensed in response to heat shock (Petesch and Lis, 2008). As expected, upon heat shock of S2 cells, we detected a reduction in nucleosome occupancy throughout the hsp70 transcriptional start site (TSS) and gene body (Fig. S4 D), as indicated by a reduction of normalized qPCR signal in the digested mononucleosomal fraction (as described in Petesch and Lis, 2008). The detected heat shock–induced difference in nucleosomal occupancy of hsp70 supports the validity of this assay to measure levels of chromatin decondensation.
To test if Elys regulates nucleosome compaction levels at endogenous targets Hph and B52, we used the MNase-qPCR assay on S2 cells treated with control or Elys RNAi (Fig. S4 E). We found an increase in the occupancy of multiple nucleosomes throughout the TSS and gene body of Hph and B52 upon dsElys RNAi treatment relative to dsWhite control (Fig. 7, A and C; and Fig. S4 E), suggesting an increase in chromatin compaction upon loss of Elys. To determine if reduction of Elys levels also affected gene expression, we tested transcript levels by RT-qPCR and found a significant reduction in the expression of both transcript isoforms of Hph (Fig. 7 B). This result supports the physiological relevance of Elys chromatin binding and regulation. Interestingly, expression of B52 remained unaffected in Elys RNAi conditions (Fig. 7 D), despite increased nucleosomal occupancy that we observed in the same conditions (Fig. 7 C). We postulate that B52 may be regulated in a different manner from Hph, such that the increase in chromatin condensation, caused by Elys depletion, is not sufficient to result in a significant down-regulation of expression of B52. However, the fact that Elys consistently affects chromatin compaction, regardless of its effect on expression again suggests that chromatin decondensation is a primary chromatin-associated function of certain Nups such as Elys.
Figure 7.
Elys regulates levels of chromatin compaction and gene expression at endogenous gene targets. (A) Graph displaying nucleosome occupancy levels along a region spanning the first ∼600 bp downstream and ∼200 bp upstream from the TSS of Hph transcripts RA and RB (TSS marked as bp “0”). Nucleosome occupancy was measured by the ratio of digested to undigested chromatin (quantified by qPCR), retrieved following MNase digestion of genomic DNA from Drosophila S2 cells treated with control dsWhite or dsElys RNAi. A schematic of corresponding regions of Hph RA and RB transcripts is shown below the graph. Error bars represent SEMs. Means and error bars were obtained from three independent biological replicates here and in B–D. (B) Expression data for HA and HB isoforms of Drosophila gene Hph, measured by RT-qPCR in Drosophila S2 cultured cells treated with control dsWhite or dsElys RNAi. Error bars represent SDs. (C) Graph displaying nucleosome occupancy levels along a region spanning ∼1000 bp downstream of B52 TSS (TSS marked as bp “0”). Nucleosome occupancy was measured by the ratio of digested to undigested chromatin (quantified by qPCR), retrieved following MNase digestion of genomic DNA from Drosophila S2 cells treated with control dsWhite or dsElys RNAi. A schematic of corresponding regions of B52 transcript is shown below the graph. Error bars represent SEMs. (D) Expression data using primers against two regions of Drosophila gene B52, measured by RT-qPCR in Drosophila S2 cultured cells treated with control dsWhite or dsElys RNAi. The two target primer locations correspond to different locations within the B52 gene region. Error bars represent SDs. (E) Model for chromatin state regulation by Nups, whereby binding of Elys and Sec13 to chromatin recruit GAF and the chromatin-remodeling complex PBAP, which promote chromatin decondensation/opening. Under the proper developmental context, this may allow for transcription factors to access target genetic elements, promote RNAP II binding and activation, and contribute to subsequent downstream gene expression at Nup target genes.
Discussion
The specific roles of different nuclear pore components in regulation of chromatin and gene expression remain poorly characterized. Our presented findings, combined with previous findings in the field demonstrating functional roles for Nups in regulating gene expression (Capelson et al., 2010; Kalverda et al., 2010; Light et al., 2010, 2013; Pascual-Garcia et al., 2014; Franks et al., 2017; Toda et al., 2017), lead to a model whereby certain Nups primarily influence chromatin state, which in turn can affect downstream gene expression (Fig. 7 E). We propose that chromatin-bound Nups, such as Elys and Sec13, recruit factors associated with formation of open chromatin, specifically GAF and components of PBAP. This results in a permissive, open-chromatin state, which, in the right cellular contexts, may allow for binding of appropriate transcription factors, for RNAP II recruitment, and subsequently in an increase in downstream gene expression (Fig. 7 E). Together, our results and model suggest a specific chromatin-decondensing function of certain Nups, particularly Elys, as an early step in the process of gene activation.
Our article provides evidence that Nups facilitate chromatin decondensation. The resulting “holes” in DNA stain that appear upon Nup tethering (Fig. 1 A) and the associated loss of IF signal from antibodies against both core histone H3 (Fig. 2, A and B) and histone modifications (Figs. 2 B and S2 A) favor this notion. This interpretation is further supported by the observed recruitment and functional involvement of the chromatin-remodeling PBAP complex (Figs. 5 and S3 B) and by additional biochemical data showing global (Fig. 6 B) and gene-specific (Fig. 7, A and B) defects in nucleosome occupancy upon Elys depletion. Furthermore, the robust biochemical interaction between Nups and components of PBAP (Fig. 6 A) and the correlation between the amount of Brm recruitment by Nups and the level of observed decondensation at lacO 96C (Figs. 1 E and 5 A) further suggest that Nups have the capacity to promote chromatin decondensation. One interesting outstanding question is whether these Nup-induced changes in chromatin structure can occur entirely de novo (or rapidly after Nup binding) or require the process of chromatin assembly during replication to take effect. Our experiments have not differentiated between these possibilities. Further experiments, perhaps in blocking replication and assaying for similar Nup functions, could differentiate between these mechanisms further. Regardless, our findings strongly support the function of Nups in regulating compaction states of chromatin, while the particular cell cycle stage and the dynamic time frame at which this process takes place remain to be elucidated.
As transcription and chromatin decompaction are intimately intertwined, we were interested to know if our Nup-induced changes in chromatin were primary or secondary to transcriptional regulation. We observed increased transcription of the gene directly downstream from the lacO 96C integration, dan, upon tethering of Sec13, which also promoted decondensation here. However, since Brm recruitment and chromatin decondensation appear to be much more robustly detected (Figs. 1 E and 5 A) than the presence of RNAP II (Fig. 2 C) upon Sec13 tethering, we believe that decondensation is likely the primary effect of Sec13 tethering, and increased gene expression a secondary consequence. This is supported by the fact that Nup62 is able to induce a small amount of detectable decondensation at 96C lacO (Fig. 1 E), associated with low-level recruitment of Elys (Fig. 4 B) and Brm (Fig. 5 A), but does not result in significant levels of RNAP II recruitment (Fig. 2 C). Further evidence that the primary effect of Nup targeting is decondensation rather than transcriptional activation is the increased nucleosome occupancy at both Hph and B52 genes upon Elys knockdown (Fig. 7, A and C), but a transcriptional change is detected only at Hph (Fig. 7, B and D). The differential effect of Elys depletion on Hph and B52 transcription again suggests that the primary chromatin-associated role of certain Nups is to facilitate the step of chromatin decondensation.
Although we found tethering of Sec13 to elicit chromatin decondensation in the ectopic context, our data suggest that Elys may be the Nup primarily responsible for facilitating decondensation. As discussed above, there is a striking correlation between levels of Elys recruitment and level of decondensation at multiple lacO loci (Fig. 4, B and C; and Fig. 1, D and E), and endogenous Elys appears to interact much more robustly with components of PBAP in S2 cells than Sec13 (Fig. 6 A). Significantly, Elys depletion shows a defect in global genomic MNase digestion, whereas Sec13 depletion does not (Fig. 6 B). The latter experiment also suggests that the role of Elys in chromatin decondensation is independent of NPC integrity, as both Elys and Sec13 (which is a core component of the Nup107–Nup160 complex) are required for nuclear pore assembly (Walther et al., 2003; Rasala et al., 2006; Franz et al., 2007). Therefore, a lack of phenotype of Sec13 RNAi in the MNase assay suggests that the observed reduction in nucleosomal accessibility in Elys RNAi conditions does not stem from a defect in NPC assembly. This conclusion is supported by the previously published observation that inhibiting transport capabilities of the NPC with WGA treatment does not lead to chromatin decondensation defects (Aze et al., 2017). We further hypothesize that since Elys exhibits a particularly robust genome-wide binding (Pascual-Garcia et al., 2017) while Sec13 appears to bind fewer loci (Capelson et al., 2010), Elys has a stronger and more detectable effect on global chromatin decompaction. It remains to be determined whether Sec13 and Elys share a subset of target genes, and whether chromatin-bound Sec13 co-functions with Elys in chromatin decompaction of such targets.
The data presented here provide functional and mechanistic evidence for the long-standing visual correlation between NPCs and open chromatin and validates the hypothesized relationship between them. Interestingly, previous genetic and proteomic experiments have reported interactions between the Caenorhabditis elegans homologue of Elys, MEL-28, and chromatin-remodeling complexes, including the SWI/SNF complex subunit SWSN-2.2 (Fernandez et al., 2014; Ertl et al., 2016), suggesting an evolutionarily conserved role for Elys in regulating chromatin state. Furthermore, genetic and physical interactions between yeast NPC components and the chromatin-remodeling RSC complex have also been reported (Titus et al., 2010; Van de Vosse et al., 2013). Elys is known to bind condensed postmitotic chromatin to nucleate NPC assembly during nuclear envelope reformation (Franz et al., 2007), and recent work has reported a defect in global postmitotic chromatin decompaction associated with depletion of Elys from chromatin (Aze et al., 2017). Thus, an intriguing possibility is that in addition to NPC assembly, postmitotic chromatin binding of Elys may also play a role in postmitotic chromatin decompaction through mechanisms similar to those we have described here. A role for Nups in facilitating the formation or maintenance of open chromatin is also consistent with the evolutionarily conserved phenomenon of viral genome integration into open/active chromatin regions that are associated with NPCs (Lelek et al., 2015; Marini et al., 2015; Manhas et al., 2018). Finally, the interaction of Nups with developmentally critical GAF and PBAP suggests that this relationship may be relevant to the establishment of tissue-specific open chromatin regions or the global genome decompaction during organismal development. It is possible that the potential role of Elys and possibly other Nups in postmitotic chromatin decondensation has extended to regulation of chromatin structure in the context of interphase transcription, thus contributing to regulation of developmental transcriptional programs.
Materials and methods
Cloning, transgenic line generation, and protein verification
Gateway cloning was used to add the LacI sequence (NCBI Escherichia coli GeneID 945007; available from GenBank under accession no. NC_000913.3), missing the last eight amino acids that represent the tetramerization domain, on the N terminus of full-length Nup62 or Sec13 within a pTWM Gateway vector containing a C-terminal myc tag and N-terminal UAS regulatory sequence. These were sent to BestGene Inc. for embryo injection for random p-element–mediated genomic integration. Lines were verified by homogenizing five larvae per genotype in Laemmli buffer to generate protein extracts, running the protein extracts out by SDS-PAGE, transferring the protein onto nitrocellulose membranes, and Western blotting the resulting membranes with α-LacI antibody (Fig. S1).
Drosophila stocks and genetics
Drosophila were raised at 22°C on standard molasses fly food. Stocks with genomically integrated lacO arrays are as follows: lacO-96C (line P11.3 from Li et al., 2003), lacO-60F (Bloomington Drosophila Stock Center [BDSC] #25371; generated by the John W. Sedat laboratory, University of California, San Francisco, San Francisco, CA), and lacO-4D5 (from Danzer and Wallrath, 2004). Crosses for larval salivary gland IF were made using females from generated stocks containing lacO-4D5 and driver Nub-Gal4 (BDSC #42699), lacO-60F and driver Sgs3-Gal4 (BDSC #6870), or lacO-96C and driver Nub-Gal4, crossed to homozygous males from UAS-LacI-Nup fusion lines or UAS-LacI-GFP (Danzer and Wallrath, 2004). Brm RNAi KD line is BDSC #35211. Larvae were raised in undercrowded conditions and dissected at later wandering third instar stage, where larvae are minimally moving but anterior spiracles have not yet protruded.
Polytene chromosome squashing, immunostaining, and fluorescence imaging
Salivary glands were dissected from wandering third instar D. melanogaster larvae in 0.1% PBS with Tween 20 (PBST), fixed in 2% PFA/45% acetic acid for 1 min at RT, squashed in a drop of 45% acetic acid between a Sigmacote (SL2; Sigma-Aldrich) coverslip and a poly-L-lysinated slide (Polysciences 22247) with a rubber hammer, and snap-frozen in liquid nitrogen; coverslips were flipped off, and slides were stored for <1 h in 0.1% PBST in a coplin jar before blocking in 3% BSA PBST for 30 min at RT and incubated overnight at 4°C in 30 µl in a blocking solution containing primary antibodies under a coverslip in a humid chamber. The following day they were washed three times for 10 min each in PBST, stained with secondary antibodies in blocking solution for 1 h at RT in the dark, and then washed three times for 10 min each time again before treatment with 10 μg/ml Hoechst stain in PBS for 2 min followed by a 5-min PBS wash before mounting in ProLong Gold Antifade (P36930; ThermoFisher), sealing with nail polish, and storage at 4°C. Slides were imaged within 1 wk of fixation. Widefield fluorescence imaging was conducted at room temperature on a Leica DM6000 Microscope with PL APO 100×/1.40-0.70 Oil objective using Type F Immersion Oil Leica 11513859, DFC365 FX Camera, and Leica LAS-X 3.3 Software. Confocal imaging was conducted at room temperature on a Leica TCS SP8 Confocal using PL APO 63×/1.40 Oil objective, 4× Zoom, Type F Immersion Oil Leica 11513859, and Leica Software LAS-X 3.3. The fluorochromes used are listed in Antibodies for IF. A minimum of 3 animals and, on average, 10–15 lacO sites per animal were imaged and analyzed for all experiments, with the exception of squashes with lacO-60F due to limitations in the ability to reliably localize sufficient LacI protein levels bound to lacO, possibly due to the repetitive nature of this locus in the subtelomeric chromatin being frequently under-replicated.
Antibodies for IF
The primary antibodies and dilutions used were as follows: GFP #1020 from Aves Labs Inc. at 1:500; LacI #600-401-B04S from Rockland Inc. at 1:100; Myc 9E10/sc-40X from Santa Cruz Biotechnology at 1:100; mAb414 (NPC marker) #902901 from Biolegend at 1:20; H3 #39763 from Active Motif at 1:100; HOAP from the Yikang S. Rong Lab (Sun Yat-sen University, Guangzhou, China) at 1:100; H3K27ac #39135 from Active Motif at 1:100; H3K4me2 #39141 from Active Motif at 1:100; H5 (Ser2ph Pol II) #920204 from Biolegend at 1:20; Mtor #12F10 from the Developmental Studies Hybridoma Bank at 1:30; Brm, Bap60, and polybromo from the Susumu Hirose Lab (National Institute of Genetics, Mishima, Shizuoka, Japan) at 1:100; GAF from the Julia Zeitlinger Lab (Stowers Institute for Medical Research, Kansas City, MO), at 1:50; CTCF from the Victor Corces Lab (Emory University, Atlanta, GA) at 1:100; Nup107 #29864 from the Capelson Lab (University of Pennsylvania, Philadelphia, PA) at 1:100; Nup93 #2648 at 1:100 and Elys at 1:50, both from the Capelson Lab (Pascual-Garcia et al., 2017); and Hoechst DNA stain (H3570; ThermoFisher) at 1:1,000. Fluorescently conjugated secondary antibodies were as follows: ThermoFisher Alexa Fluor conjugates of goat anti-mouse, anti-rabbit, and anti-guinea pig to 488 and 568.
H3 alternative fixation conditions for polytene squashes
Polytene chromosome squashes for use with the H3 antibody required an alternative fixation protocol to prevent extraction of histones from chromatin, which replaced standard fixation of glands with a 30 s fix in 2% PFA, followed by 2 min in 2% PFA/45% acetic acid, and a final placement into a drop of 45% acetic acid during squashing, all at RT. After flash freezing in liquid nitrogen, slides were kept at −20°C in 70% ethanol for ≥30 min before two quick rinses in PBST and the standard subsequent blocking and staining protocol.
Polytene chromosome nuclei semi-squashes
Semi-squashes used to better preserve nuclear shape to verify rim staining of LacI-Nup fusions (Fig. S1) use an identical protocol as full squashes but instead with a 2-min fixation in 8% acetic acid/2% PFA and a 2% PFA droplet used on the coverslip, at which point the coverslip is not hammered but is gently moved ∼1 mm in each direction two times before freezing. Antibodies and dilutions are listed in Table S1.
PCC analysis
Intensity correlation analysis was performed to determine the extent to which a given LacI fusion protein (the “tester”) resulted in enrichment or depletion of components of chromatin-modifying complexes or other factors (the “targets”). Each image consisted of three channels representing Hoechst and the IF signals of the tester and target. To select pixels for inclusion in the correlation calculation, image segmentation was performed on the Hoechst and tester images using custom MATLAB software. First, manual input was used to select a candidate threshold from the Hoechst DNA image, followed by balanced histogram thresholding of the tester. Further manual input was used to refine the tester- and Hoechst-based masks to ensure that (1) the majority of pixels included in the correlation calculation contained nonbackground levels of tester signal and that (2) these signals were localized to the chromosome. Values reported are linear PCCs calculated using target-tester value pairs for all pixels found in the joint Hoechst-tester mask. In cases of measuring chromatin decondensation, the Hoechst channel was used as the target as well. Statistical significance was determined by a one-way ANOVA test with Tukey’s multiple comparisons post-test when three genotypes are compared and an unpaired t test when only two genotypes are compared.
HOAP area quantification
Using ImageJ, red HOAP capping signals at the telomere of chromosome 2R, designated by the presence of LacI-fusion protein signal at adjacent lacO-60F, were manually traced, and the areas were measured and compared for each condition. Statistical significance was determined by a one-way ANOVA test with Tukey’s multiple comparisons post-test.
3D FISH in intact salivary glands
Inverted larval heads (removing fat, gut, and heart but preserving brain, discs, and glands) were dissected in cold PBS, collected on ice, and fixed using 200 μl 4% PFA/0.5% IGEPAL/PBS + 600 μl heptane, hand-shaken vigorously, and incubated for 10 min on a nutator. Larval heads were then washed in PBST three times for 5 min each, rinsed three times in 2XSSCT, transferred to 20% formamide in 2XSSCT for 10 min at RT, transferred to 50% formamide for 10 min at RT, and then transferred to 50% formamide for 3–5 h at 37°C on a rocker in a hybridization oven. Heads were then incubated in 100 µl hybridization buffer (2XSSCT/10% dextran sulfate/50% formamide) + 200 ng lacO probe (sequence listed in Table S1) for 30 min at 80°C before overnight incubation with rocking in a hybridization oven at 37°C. After probe incubation, heads were washed two times in 50% formamide for 30 min at 37°C, washed in 20% formamide for 10 min at RT, rinsed four times in 2XSSCT, stained with 10 μg/ml Hoechst in 2XSSCT for 5 min, washed for 5 min in 2XSSCT and for 10 min in 2XSSC, after which glands were dissected from heads in 2XSSC, gently mounted in nonhardening VectaShield antifade (H-1000; Vector-Labs), and stored upside down in a slide box with raised slots to prevent nuclei flattening until imaging using 3D confocal microscopy.
Brm reduction quantification
Using ImageJ, green LacI-Sec13 protein fusion bands at each lacO-96C site were manually traced, and the corresponding mean intensity value of the red Brm fluorescence signal under those bands was measured. The mean fluorescence intensity of the nearby Brm control band, which is located in the interband of nearby 96 D and was observed to remain unchanged between preparations at this stage in development, was also measured. A ratio between each lacO/control band was generated and plotted for control and Brm KD conditions. Statistical significance was determined by unpaired t test.
CoIP and Western blotting
S2 cells were harvested and washed twice in PBS. 3 × 107 cells were resuspended in 250 μl of high-salt buffer (10 mM Tris-HCL, pH 7.4; 400 mM NaCl; 1% Triton X-100; 2 mM EGTA; 1 mM MgCl2; 1 mM DTT; and C0mplete EDTA-free Tablet [1 per 10 ml]; Sigma-Aldrich 11873580001) and Pierce Nuclease (1:500) for 45 min at 4°C. The sample was then sonicated three times for 10 s each on setting 2 of a Fischer Sonic Dismembrator Model 100, resting 10 s on ice between sonications. The sample was spun down at 10,000 relative centrifugal force for 10 min, and 500 μl of no-salt buffer (10 mM Tris-HCL, pH 7.4; 2 mM EDTA; and 1 mM DTT) was added. 6 μl of antibody was added to the lysate mixture and incubated overnight on a rotator at 4°C. 30 μl of Dynabeads (Life Technologies) were washed in blocking buffer (0.3% BSA in PBS) and blocked for 30 min. Beads were washed in no-salt buffer once, added to the antibody/lysate mixture, and incubated on a rotator for 3 h at 4°C. After incubating, beads were washed five times in wash buffer (1:3 high-salt:no-salt), eluted in 1× Laemmli buffer, run on SDS-PAGE gel, transferred to membrane, and blotted against indicated antibodies.
Cell culture and RNA interference
Drosophila S2 cells were grown in Schneider’s medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO) and antibiotics. dsRNA against Elys or White genes were generated from PCR templates of fly genomic DNA using specific T7 primers (Elys-F 5′-TAATACGACTCACTATAGGGAGAGCACGTATCTTCGCATCAGA-3′; Elys-R 5′-TAATACGACTCACTATAGGGAGAGACAAGGACGCTTATTGGGA-3′; White-F 5′-TAATACGACTCACTATAGGGAGAGATCCTGGCTGTCGGTGCTCA-3′; White-R 5′-TAATACGACTCACTATAGGGAGAGATCATCGGATAGGCAATCGC-3′) and Sec13 primer set 2 from Capelson et al. (2010). dsRNAs were synthesized using a Megascript T7 kit (Ambion) following the manufacturer’s instructions. S2 cells were seeded at 15 × 106 cells per plate in a 10-cm dish plate, treated with 10 μg of specific dsRNA per 106 cells every 48 h, and harvested after 6 d of treatment.
RNA extraction and RT-qPCR expression analysis
Total RNA was isolated using Trizol (Ambion) from salivary glands vortexed at 4°C for 2 h or from S2 cell pellets vortexed for 30 min, extracted with ethanol precipitation, and subsequently purified with PureLink RNA Kit columns (Invitrogen). 1 µg of the extracted RNA was used for first-strand cDNA synthesis using a one-step RT-PCR kit (Qiagen). To measure mRNA levels, real-time qPCRs were performed on resulting cDNA using gene-specific primers, as listed in Table S1.
Global MNase digestion assay
MNase accessibility assays were performed on equal amounts of collected dsRNA-treated S2 cells (described above). Cells were incubated for 10 min on ice with buffer A (15 mM Tris, pH 7.4; 60 mM KCl; 15 mM NaCl; 5 mM MgCl2; 300 mM sucrose; and 0.1% IGEPAL) and treated to 10 strokes using a Dounce homogenizer. Lysate was centrifuged and washed once with buffer A without detergent. Nuclei were then resuspended in MNase buffer (15 mM Tris, pH 7.4; 60 mM KCl; 15 mM NaCl; 3 mM CaCl2; and 200 mM sucrose) and digested at 37°C with 1 U MNase (#2910A; Takara). The reaction was stopped by adding 0.15 volumes of Stop solution (4% SDS and 100 mM EDTA). RNA and proteins were digested with 70 μg of RNase A for 1 h at 37°C followed by 70 μg of freshly made proteinase K for 2 h at 55°C. Digested DNA was purified with phenol chloroform extraction followed by ethanol precipitation. Finally, DNA was resuspended in TE (10 mM Tris, pH 8.0; and 1 mM EDTA) and analyzed on a 1.7% agarose gel stained with ethidium bromide.
Heat shock of S2 cells
For heat shock treatment, we followed Petesch and Lis (2008). The media volume of S2 cells growing at 25°C in a 10-cm dish plate was adjusted to 7.5 ml. To heat shock the cells, we added 7.5 ml of media that was prewarmed at 48°C and incubated the cells for 3 min at 37°C. Heat shock treatment was stopped by supplementing the media with 5 ml of 4C media, and cells were then immediately fixed for the downstream MNase-qPCR procedure.
MNAse-qPCR
MNase-qPCR experiments were performed as described previously (Infante et al., 2012), with some modifications. S2 cells were fixed with 1% formaldehyde for 10 min with gentle rotation. Fixation was quenched adding glycine to a final concentration of 125 mM, and then cells were washed twice with cold PBS. Cells were then resuspended in 3 ml of buffer A (10 mM Tris, pH 8.0; 3 mM CaCl2; 2 mM magnesium acetate; 300 mM sucrose; and 0.5 mM DTT) + 1% Triton X-100, and lyses were promoted with five passes through a 25-G needle. Lysates were washed twice with buffer A and once with buffer D (50 mM Tris, pH 8.0; 5 mM magnesium acetate; 5 mM DTT; and 25% glycerol). The nuclei were then resuspended in 200 μl MNase buffer (15 mM Tris, pH 7.4; 60 mM KCl; 15 mM NaCl; 2 mM CaCl2; 0.5 mM DTT; and 25% glycerol) and incubated for 10 min at 37°C before the addition of 120 U MNase (#2910A; Takara). Digestion was conducted at 37°C for 30 min. For each of the conditions, we ran in parallel an undigested sample with no MNase enzyme that was used for normalization purposes during qPCR analysis. MNase digestion was stopped by adding SDS and EDTA to a final concentration of 0.5% and 12.5 mM, respectively. Reverse cross-linking was achieved by incubating samples at 65°C overnight, and RNA and proteins were then digested with 70 μg RNase A and proteinase K. Finally, DNA was recovered with phenol-chloroform extraction followed by ethanol precipitation. To enrich for mono-nucleosomes, digested samples were run in an agarose gel, and mono-nucleosomes were gel-purified following standard procedures. Undigested and mono-nucleosome–enriched DNA was then quantified using a Qubit fluorometer following the commercial protocol.
Subsequent qPCR analysis is also detailed in Infante et al. (2012). The primers used are listed in Table S1. We determined the relative amount of each primer set in the undigested genomic DNA and the gel-purified mono-nucleosome DNA. The relative protection value was then calculated for each amplicon, which corresponded to the fold enrichment of the target sequence in the mono-nucleosomal DNA sample over the undigested DNA sample. Finally, we normalized the relative protection values for each amplicon to differences in DNA concentration among different samples.
ChIP-qPCR
Cells were cross-linked with 1% methanol-free formaldehyde and quenched with 0.125 mM glycine. Cells were then harvested and washed with PBS + 0.2 mM PMSF. Cells were then treated with ChIP buffer I (50 mM HEPES, pH 7.6; 140 mM NaCl; 1 mM EDTA; 0.5 mM EGTA; 10% glycerol; 0.5% IGEPAL; 0.25% Triton X-100; and C0mplete protease inhibitors (11836170001; Sigma-Aldrich), incubated on a rotator at 4°C, and spun down at 4°C. Pellets were resuspended in ChIP buffer II (20 mM Tris, pH 8.0; 200 mM NaCl; 1 mM EDTA, pH 8.0; 0.5 mM EGTA, pH 8.0; and C0mplete protease inhibitors), incubated on a rotator at 4°C, and spun down at 4°C. Pellets were resuspended in ChIP buffer III (20 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5% Sarkosyl, 0.1% sodium deoxycholate, and C0mplete protease inhibitors) and sonicated in a S220 Covaris (peak power: 140; duty ratio: 5; cycles: 200) for 15 min. Samples were transferred into 1.5-ml Lo-bind tubes, Triton X-100 to 1% at final volume was added, and samples were spun down at maximum speed for 10 min at 4°C. Supernatants were then quantified using a Bradford assay. Immunoprecipitates were set up with 200 μg of protein (12 μl of Elys antibody and 2 μl of IgG antibody) and dilution buffer (20 mM Tris, 100 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA) in a 1:2 ratio of lysate:dilution buffer. Immunoprecipitates were incubated on a rotator overnight at 4°C and 10% input and verification samples were stored at −80°C. 40 μl of Dynabeads per immunoprecipitate were washed and then blocked in 0.3% BSA in PBS on a rotator overnight at 4°C. Beads were then washed twice in dilution buffer and added to the immunoprecipitates and incubated on a rotator at 4°C. After incubation, beads were washed in low-salt buffer (20 mM Tris-HCl, pH 8; 150 mM NaCl; 2 mM EDTA; 0.1% SDS; and 1% TritonX-100), high-salt buffer (20 mM Tris-HCl, pH 8; 500 mM NaCl; 2 mM EDTA; 0.1% SDS; and 1% Triton X-100), and LiCl buffer (10 mM Tris-HCl, pH 8; 250 mM LiCl; 1% IGEPAL; 1% sodium deoxycholate; and 1 mM EDTA) once followed by TE50 (10 mM Tris-HCl, pH 8.0; 50 mM NaCl; and 1 mM EDTA) twice. Beads were resuspended in elution buffer (100 mM NaHCO3 and 1% SDS) and eluted at 65°C at 600 rpm for 30 min. Samples (immunoprecipitates and inputs) were de–cross-linked at 65°C. After de–cross-linking, equal-volume TE (10 mM Tris-HCl, pH 8.0; and 1 mM EDTA) was added to samples, and 0.2 mg/ml final concentration RNase A was added and incubated at 37°C, followed by addition of 0.2 mg/ml final concentration proteinase K and incubation at 55°C. 1× sample volume of phenol/chloroform/isoamyl alcohol was added, and samples were incubated at RT and then spun down at maximum speed. 1× sample volume of chloroform/isoamyl alcohol was added to the aqueous layer, and samples were incubated at RT and then spun down at maximum speed. 0.1× sample volume of sodium acetate (pH 5.2, final concentration of 0.3 M), 1.5 µl glycogen (stock 20 mg/ml; Roche), and 2.5× sample volume of cold 100% ethanol was added to the aqueous layer, and samples were mixed and incubated at −20°C. Samples were then spun down at maximum speed, and the DNA pellet was washed with 70% cold ethanol, spun down at maximum speed, and then air-dried until all ethanol was removed. DNA pellets were then resuspended in TE buffer and used for downstream qPCR analysis, using primers listed in Table S1.
Online supplemental material
Fig. S1 is related to Fig. 1. Fig. S2 is related to Figs. 2 and 4. Fig. S3 is related to Fig. 5. Fig. S4 is related to Figs. 6 and 7. Table S1 contains sequences of all the primers used in the manuscript for the RNAi/dsRNA, RT-qPCR, ChIP-qPCR, and MNAse-qPCR applications, as well as FISH oligo probe sequence.
Supplementary Material
Acknowledgments
We are exceedingly grateful to Susumu Hirose for his gift of antibodies to the PBAP complex, and Lori Wallrath (University of Iowa, Iowa City, IA) for great effort in tracking down the sequencing results to locate the lacO-96C (P11.3) insertion site. We also thank Julia Zeitlinger, Victor Corces, Mia Levine (University of Pennsylvania, Philadelphia, PA), and the Developmental Studies Hybridoma Bank for providing critical antibodies. We thank the BDSC, Kristen Johansen (Iowa State University, Ames, IA), and Lori Wallrath for key fly stocks; Eric Joyce laboratory (University of Pennsylvania, Philadelphia, PA) members for help with oligoprobe design; and the University of Pennsylvania CDB Microscopy Core Facility for confocal microscope use.
T.M. Kuhn is supported by National Institutes of Health grant T32 HD083185. M. Capelson is supported by the American Cancer Society Research Scholar Grant RSG-15-159-01-CSM and by National Institutes of Health grant R01GM124143.
The authors declare no competing financial interests.
Author contributions: T.M. Kuhn and M. Capelson conceived the ideas and experimental design, interpreted results, and wrote the manuscript. T.M. Kuhn planned and executed the experiments and data analysis, with the exception of experiments in Fig. S2 A; Fig. 6 A; and Fig. S4, B and C, which were executed by A. Gozalo, and Fig. 6, B and C; Fig. S4, A, D, and E; and Fig. 7, A–D, which were executed by P. Pascual-Garcia. The method of image quantification analysis for protein recruitment was conceived by S.C. Little and T.M. Kuhn, developed by S.C. Little, and executed by T.M. Kuhn.
References
- Aze A., Fragkos M., Bocquet S., Cau J., and Méchali M.. 2017. RNAs coordinate nuclear envelope assembly and DNA replication through ELYS recruitment to chromatin. Nat. Commun. 8:2130 10.1038/s41467-017-02180-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc. Natl. Acad. Sci. USA. 82:8527–8529. 10.1073/pnas.82.24.8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner D.G., Cajigas I., Fondufe-Mittendorf Y., Ahmed S., Lee P.C., Widom J., and Brickner J.H.. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal G.G., Genovesio A., Rodriguez-Navarro S., Zimmer C., Gadal O., Lesne A., Buc H., Feuerbach-Fournier F., Olivo-Marin J.C., Hurt E.C., and Nehrbass U.. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 441:770–773. 10.1038/nature04752 [DOI] [PubMed] [Google Scholar]
- Capelson M., and Hetzer M.W.. 2009. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 10:697–705. 10.1038/embor.2009.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., and Hetzer M.W.. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383. 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari J.M., Brown C.R., Komili S., West J., Hieronymus H., and Silver P.A.. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 117:427–439. 10.1016/S0092-8674(04)00448-9 [DOI] [PubMed] [Google Scholar]
- Cenci G., Siriaco G., Raffa G.D., Kellum R., and Gatti M.. 2003. The Drosophila HOAP protein is required for telomere capping. Nat. Cell Biol. 5:82–84. 10.1038/ncb902 [DOI] [PubMed] [Google Scholar]
- D’Urso A., and Brickner J.H.. 2017. Epigenetic transcriptional memory. Curr. Genet. 63:435–439. 10.1007/s00294-016-0661-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso A., Takahashi Y.H., Xiong B., Marone J., Coukos R., Randise-Hinchliff C., Wang J.P., Shilatifard A., and Brickner J.H.. 2016. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. eLife. 5:e16691 10.7554/eLife.16691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danzer J.R., and Wallrath L.L.. 2004. Mechanisms of HP1-mediated gene silencing in Drosophila. Development. 131:3571–3580. 10.1242/dev.01223 [DOI] [PubMed] [Google Scholar]
- Deng H., Bao X., Cai W., Blacketer M.J., Belmont A.S., Girton J., Johansen J., and Johansen K.M.. 2008. Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development. 135:699–705. 10.1242/dev.015362 [DOI] [PubMed] [Google Scholar]
- Ertl I., Porta-de-la-Riva M., Gómez-Orte E., Rubio-Peña K., Aristizábal-Corrales D., Cornes E., Fontrodona L., Osteikoetxea X., Ayuso C., Askjaer P., et al. . 2016. Functional interplay of two paralogs encoding SWI/SNF chromatin-remodeling accessory subunits during Caenorhabditis elegans development. Genetics. 202:961–975. 10.1534/genetics.115.183533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A.G., Mis E.K., Lai A., Mauro M., Quental A., Bock C., and Piano F.. 2014. Uncovering buffered pleiotropy: a genome-scale screen for mel-28 genetic interactors in Caenorhabditis elegans. G3 (Bethesda). 4:185–196. 10.1534/g3.113.008532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks T.M., McCloskey A., Shokirev M.N., Benner C., Rathore A., and Hetzer M.W.. 2017. Nup98 recruits the Wdr82-Set1A/COMPASS complex to promoters to regulate H3K4 trimethylation in hematopoietic progenitor cells. Genes Dev. 31:2222–2234. 10.1101/gad.306753.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., and Antonin W.. 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8:165–172. 10.1038/sj.embor.7400889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda N.J., Guertin M.J., Sharma S., Danko C.G., Martins A.L., Siepel A., and Lis J.T.. 2015. GAGA factor maintains nucleosome-free regions and has a role in RNA polymerase II recruitment to promoters. PLoS Genet. 11:e1005108 10.1371/journal.pgen.1005108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M., and Sedat J.W.. 1987. Three-dimensional organization of Drosophila melanogaster interphase nuclei. II. Chromosome spatial organization and gene regulation. J. Cell Biol. 104:1471–1483. 10.1083/jcb.104.6.1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., and Hetzer M.W.. 2015. Nuclear pore proteins and the control of genome functions. Genes Dev. 29:337–349. 10.1101/gad.256495.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A., Benner C., Tyagi S., Cool J., and Hetzer M.W.. 2016. Nucleoporin-mediated regulation of cell identity genes. Genes Dev. 30:2253–2258. 10.1101/gad.287417.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante J.J., Law G.L., and Young E.T.. 2012. Analysis of nucleosome positioning using a nucleosome-scanning assay. Methods Mol. Biol. 833:63–87. 10.1007/978-1-61779-477-3_5 [DOI] [PubMed] [Google Scholar]
- Kalverda B., Pickersgill H., Shloma V.V., and Fornerod M.. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 140:360–371. 10.1016/j.cell.2010.01.011 [DOI] [PubMed] [Google Scholar]
- Lelek M., Casartelli N., Pellin D., Rizzi E., Souque P., Severgnini M., Di Serio C., Fricke T., Diaz-Griffero F., Zimmer C., et al. . 2015. Chromatin organization at the nuclear pore favours HIV replication. Nat. Commun. 6:6483 10.1038/ncomms7483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Danzer J.R., Alvarez P., Belmont A.S., and Wallrath L.L.. 2003. Effects of tethering HP1 to euchromatic regions of the Drosophila genome. Development. 130:1817–1824. 10.1242/dev.00405 [DOI] [PubMed] [Google Scholar]
- Liang Y., Franks T.M., Marchetto M.C., Gage F.H., and Hetzer M.W.. 2013. Dynamic association of NUP98 with the human genome. PLoS Genet. 9:e1003308 10.1371/journal.pgen.1003308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W.H., Brickner D.G., Brand V.R., and Brickner J.H.. 2010. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol. Cell. 40:112–125. 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W.H., Freaney J., Sood V., Thompson A., D’Urso A., Horvath C.M., and Brickner J.H.. 2013. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 11:e1001524 10.1371/journal.pbio.1001524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhas S., Ma L., and Measday V.. 2018. The yeast Ty1 retrotransposon requires components of the nuclear pore complex for transcription and genomic integration. Nucleic Acids Res. 46:3552–3578. 10.1093/nar/gky109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini B., Kertesz-Farkas A., Ali H., Lucic B., Lisek K., Manganaro L., Pongor S., Luzzati R., Recchia A., Mavilio F., et al. . 2015. Nuclear architecture dictates HIV-1 integration site selection. Nature. 521:227–231. 10.1038/nature14226 [DOI] [PubMed] [Google Scholar]
- Mohrmann L., and Verrijzer C.P.. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta. 1681:59–73. 10.1016/j.bbaexp.2004.10.005 [DOI] [PubMed] [Google Scholar]
- Nakayama T., Shimojima T., and Hirose S.. 2012. The PBAP remodeling complex is required for histone H3.3 replacement at chromatin boundaries and for boundary functions. Development. 139:4582–4590. 10.1242/dev.083246 [DOI] [PubMed] [Google Scholar]
- Ohtsuki S., and Levine M.. 1998. GAGA mediates the enhancer blocking activity of the eve promoter in the Drosophila embryo. Genes Dev. 12:3325–3330. 10.1101/gad.12.21.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollion J., Cochennec J., Loll F., Escudé C., and Boudier T.. 2013. TANGO: a generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics. 29:1840–1841. 10.1093/bioinformatics/btt276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D., Pascual-Garcia P., Dunagin M., Tudor M., Hopkins K.C., Xu J., Gold B., Raj A., Capelson M., and Cherry S.. 2014. Nup98 promotes antiviral gene expression to restrict RNA viral infection in Drosophila. Proc. Natl. Acad. Sci. USA. 111:E3890–E3899. 10.1073/pnas.1410087111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P., Jeong J., and Capelson M.. 2014. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Reports. 9:433–442. 10.1016/j.celrep.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P., Debo B., Aleman J.R., Talamas J.A., Lan Y., Nguyen N.H., Won K.J., and Capelson M.. 2017. Metazoan nuclear pores provide a scaffold for poised genes and mediate induced enhancer-promoter contacts. Mol. Cell. 66:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petesch S.J., and Lis J.T.. 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 134:74–84. 10.1016/j.cell.2008.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani H.P., and Greenleaf A.L.. 2006. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 20:2922–2936. 10.1101/gad.1477006 [DOI] [PubMed] [Google Scholar]
- Ptak C., and Wozniak R.W.. 2016. Nucleoporins and chromatin metabolism. Curr. Opin. Cell Biol. 40:153–160. 10.1016/j.ceb.2016.03.024 [DOI] [PubMed] [Google Scholar]
- Rabut G., Doye V., and Ellenberg J.. 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6:1114–1121. 10.1038/ncb1184 [DOI] [PubMed] [Google Scholar]
- Raices M., and D’Angelo M.A.. 2017. Nuclear pore complexes and regulation of gene expression. Curr. Opin. Cell Biol. 46:26–32. 10.1016/j.ceb.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M., Bukata L., Sakuma S., Borlido J., Hernandez L.S., Hart D.O., and D’Angelo M.A.. 2017. Nuclear pores regulate muscle development and maintenance by assembling a localized Mef2C complex. Dev. Cell. 41:540–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Orjalo A.V., Shen Z., Briggs S., and Forbes D.J.. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 103:17801–17806. 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood V., and Brickner J.H.. 2014. Nuclear pore interactions with the genome. Curr. Opin. Genet. Dev. 25:43–49. 10.1016/j.gde.2013.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong S.M., Wijayatilake H.D., and Proudfoot N.J.. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23:2610–2624. 10.1101/gad.1823209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus L.C., Dawson T.R., Rexer D.J., Ryan K.J., and Wente S.R.. 2010. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol. Biol. Cell. 21:1072–1087. 10.1091/mbc.e09-07-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Hsu J.Y., Linker S.B., Hu L., Schafer S.T., Mertens J., Jacinto F.V., Hetzer M.W., and Gage F.H.. 2017. Nup153 interacts with Sox2 to enable bimodal gene regulation and maintenance of neural progenitor cells. Cell Stem Cell. 21:618–634. [DOI] [PubMed] [Google Scholar]
- Tumbar T., Sudlow G., and Belmont A.S.. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354. 10.1083/jcb.145.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M., Imam N., Verma K., and Patel A.K.. 2016. Chromatin remodelers: We are the drivers!! Nucleus. 7:388–404. 10.1080/19491034.2016.1211217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Vosse D.W., Wan Y., Lapetina D.L., Chen W.M., Chiang J.H., Aitchison J.D., and Wozniak R.W.. 2013. A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell. 152:969–983. 10.1016/j.cell.2013.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas J.M., Suyama R., Kind J., Miura K., Luscombe N.M., and Akhtar A.. 2010. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 6:e1000846 10.1371/journal.pgen.1000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., et al. . 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206. 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Watson M.L. 1955. The nuclear envelope; its structure and relation to cytoplasmic membranes. J. Biophys. Biochem. Cytol. 1:257–270. 10.1083/jcb.1.3.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C., Jenness C., Kimura H., and Funabiki H.. 2014. Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 21:617–625. 10.1038/nsmb.2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.