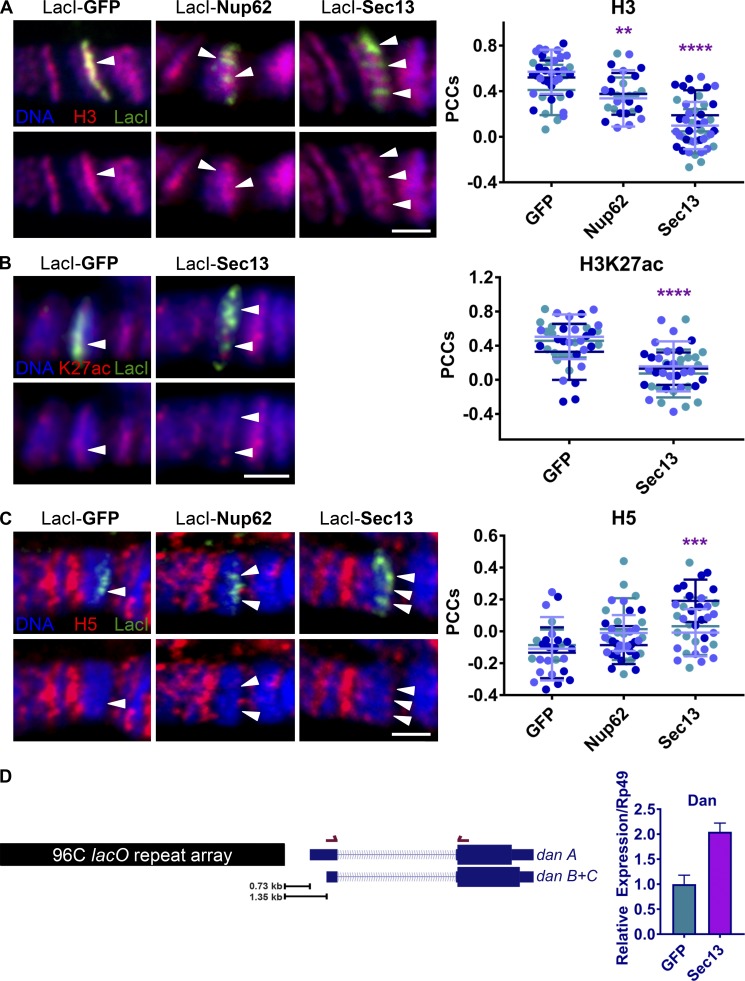
Figure 2.
Nup binding to chromatin is associated with a decrease in histone density and an increase in gene expression. (A) Confocal IF images of LacI-fusion proteins targeted to the lacO integration site on squashed polytene chromosomes at location 96C. Staining was with Hoechst (blue) and antibodies against H3 (red) and LacI (green). LacI-fusion protein expression was driven with Nubbin-Gal4. The top row shows the overlay of all three colors, whereas the bottom row shows blue and red only (here and in B and C). Arrowheads indicate locations of existing or depleted H3 under LacI signal. The scale bar is 2 µm. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 39; Nup62, n = 27; Sec13, n = 44. ****, P < 0.0001; **, P < 0.01. Error bars represent SDs. The image for control LacI-GFP is the same as the image for control LacI-GFP in Fig. 1 E, demonstrating enrichment of histone H3 and high Hoechst staining density at the same control locus. (B) Experimental conditions and strains are as in A above, but with H3K27ac antibody (red) instead of H3 and GFP or myc antibodies (green) instead of LacI due to antibody animal source constraints, and with the use of widefield microscopy. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 38; Sec13, n = 40. ****, P < 0.0001. Error bars represent SDs. (C) Experimental conditions and strains as in A above, but with antibodies against LacI (green) and CTD tail Ser2 phosphorylated RNAP II (H5, red), and with the use of widefield microscopy. Arrowheads indicate LacI signal and recruitment or lack thereof of H5. The scale bar is 2 µm. Quantification displays PCCs between red and green signal under LacI. Data are from three biological replicates (colored) from two independent experiments. GFP, n = 26; Nup62, n = 40; Sec13, n = 35. ***, P < 0.001. Error bars represent SDs. (D) Schematic of the distance between integration of the lacO repeat plasmid and the downstream isoforms of the dan gene along with location of the primer set used for RT-qPCR. Three technical replicates of each of three biological replicates (10 sets of glands per replicate) were used for quantification. Error bars represent SEMs.