Figure 3.
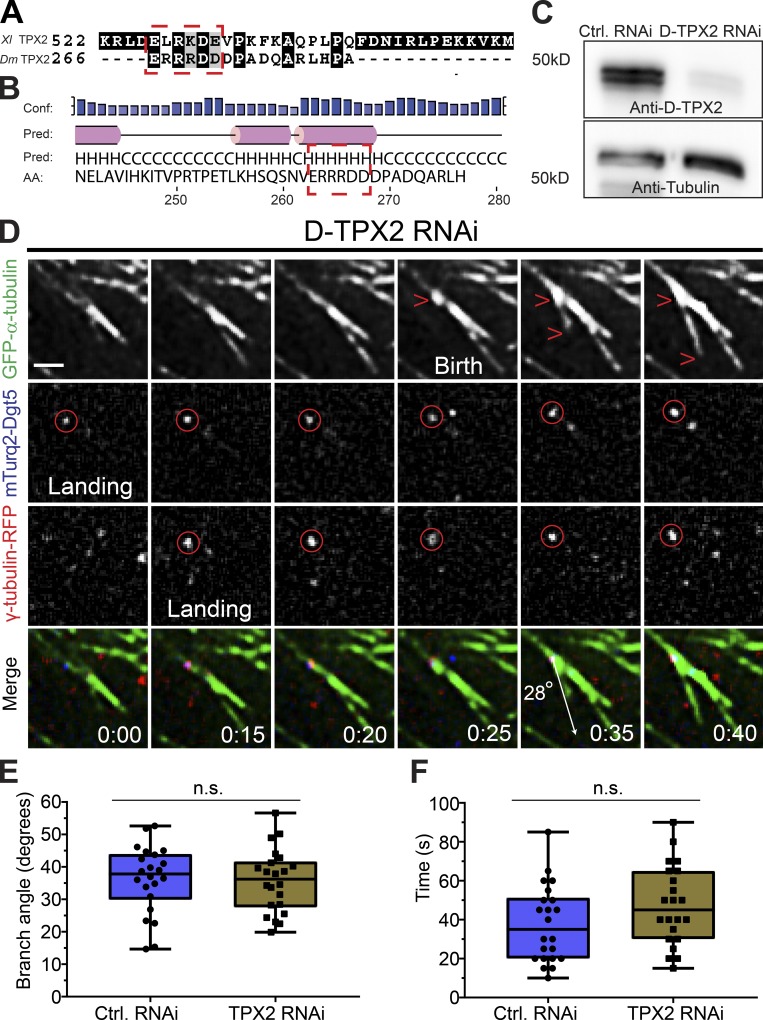
Branching MT nucleation is unaffected by D-TPX2 depletion. (A) Protein sequences of Xenopus (Xl) TPX2 and Drosophila (Dm) TPX2 (D-TPX2) were aligned using T-Coffee multiple alignment software (Notredame et al., 2000). Red dashed rectangle indicates a partially conserved CM1/γ-TuNA motif in D-TPX2. (B) Secondary structure prediction of D-TPX2 was generated using PSIPRED bioinformatics software (Buchan and Jones, 2019). Partially conserved CM1/γ-TuNA motif is expected to lie within α-helices (indicated with red dashed square). Conf, confidence; pred, predicted. (C) Western blot showing depletion of endogenous TPX2 with tubulin as a loading control (Ctrl). (D) A representative MT branching event in a D-TPX2–depleted cell coexpressing GFP-α-tubulin (green), mTurquoise2-Dgt5 (blue), and γ-tubulin-TagRFP (red). Time point 00:00 indicates landing of a Dgt5 molecule, which recruits γ-tubulin at 0:15 s. The Dgt5-γ-tubulin complex nucleates a MT branch at 0:35 s. Scale bar, 1 µm. (E) Distribution of branch angles in control and D-TPX2–depleted cells; n = 44. (F) Lifetime of branched daughter MTs in control and D-TPX2–depleted cells; n = 44. Time: min:s. Box plots indicate full range of variation (from minimum to maximum), the interquartile range, and the median. Two-tailed P values of Student’s t tests are reported; n.s., not significant or P > 0.05.