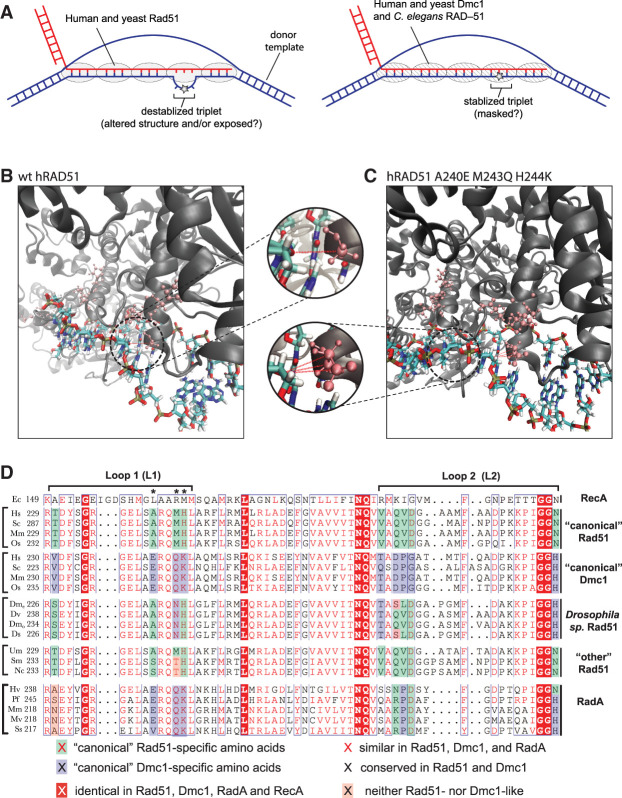
Figure 7.
Potential mechanism of mismatch stabilization and L1 conservation among different Rad51/RecA family members. (A) Model for differences between Rad51 and Dmc1 interactions involving imperfectly paired HR intermediates. (B,C) Snapshots taken from MD simulations of hRAD51 (B) and hRAD51 harboring three Dmc1 lineage-specific amino acids substitutions (A240E M243Q H244K) (C), suggesting that the Dmc1 amino acids are better positioned to contact the incoming complementary DNA strand. Insets highlight potential protein contacts (red dashed lines) with the ribose ring of the phosphate backbone. (D) Comparison of L1 and L2 sequences from E. coli RecA; “canonical” Rad51 and Dmc1 from organisms harboring both recombinases; Rad51 from four Drosophilia sp. (Drosophilia melanogaster, Drosophilia virilis, Drosophilia mojavenis, and Drosophilia simulans); Rad51 sequences designated as “other” from Ustilago maydis, Sodaria macrospora, and Neurospora crassa; and RadA from Haloferax volcanii, Pyrococcus furiousus, Methanococcus maripaludis, Methanococcus voltae, and Sulfolobus solfataricus. Asterisks denote the amino acids that contribute to mismatch stabilization for Dmc1.