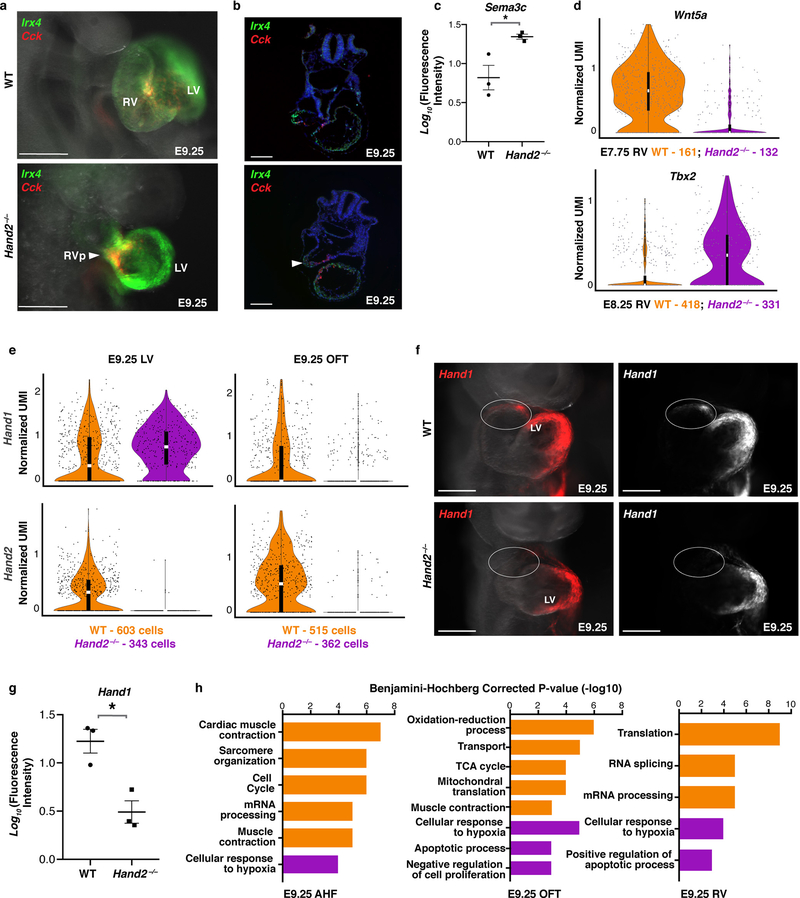
Extended Data Figure 10: Right ventricle cell migration is impaired in Hand2-null embryos.
a, Whole-mount in situ hybridization for Irx4 and Cck in right lateral view and b, transverse sections at E9.25 indicating presence of RV cells in Hand2 mutants (arrowheads). n=2 independent experiments with similar results. Scale bar, 200 μm. c, Quantification of Sema3c fluorescence signal in Fig. 4h. n=3 replicate embryos per genotype. The mean +/− s.e.m indicated. Two-tailed t-test. *P<0.05. d, Violin plots of Wnt5a and Tbx2 expression in WT and Hand2-null RV cells at E7.75 and E8.25, respectively and e, Hand1 and Hand2 expression in LV and OFT cells at E9.25. All genes in d, e have a Bonferroni correction adjusted p-value < 1×10–4 (Wilcoxon rank sum test, two-sided). Violin plot summary statistics: center white line represents median gene expression and central black rectangle spans the first to third quartile of the data distribution. Whiskers indicate value at 1.5x interquartile range above the third quartile or below the first quartile. RV, right ventricle; RVp, right ventricle progenitors; LV, left ventricle; OFT, outflow tract; AHF, anterior heart field. f, In situ hybridization for Hand1 in WT and Hand2-null embryos at E9.25 in frontal view. n=3 independent experiments with similar results. g, Quantification of Hand1 fluorescent signal in the OFT. n=3 replicated embryos per genotype. The mean +/− s.e.m is indicated. Two-tailed t-test. *P<0.05. h, GO biological process terms of differentially expressed genes in WT and Hand2-null AHF (n = 406 cells per genotype), OFT (n = 362 cells per genotype) or RV (n = 227 cells per genotype) cells at E9.25, as determined with DAVID v6.8. Significant functional enrichment was statistically determined using a modified Fisher’s exact test (EASE score) followed by Benjamini-Hochberg correction for multiple comparisons, with 0.01 as a P-value cut-off.