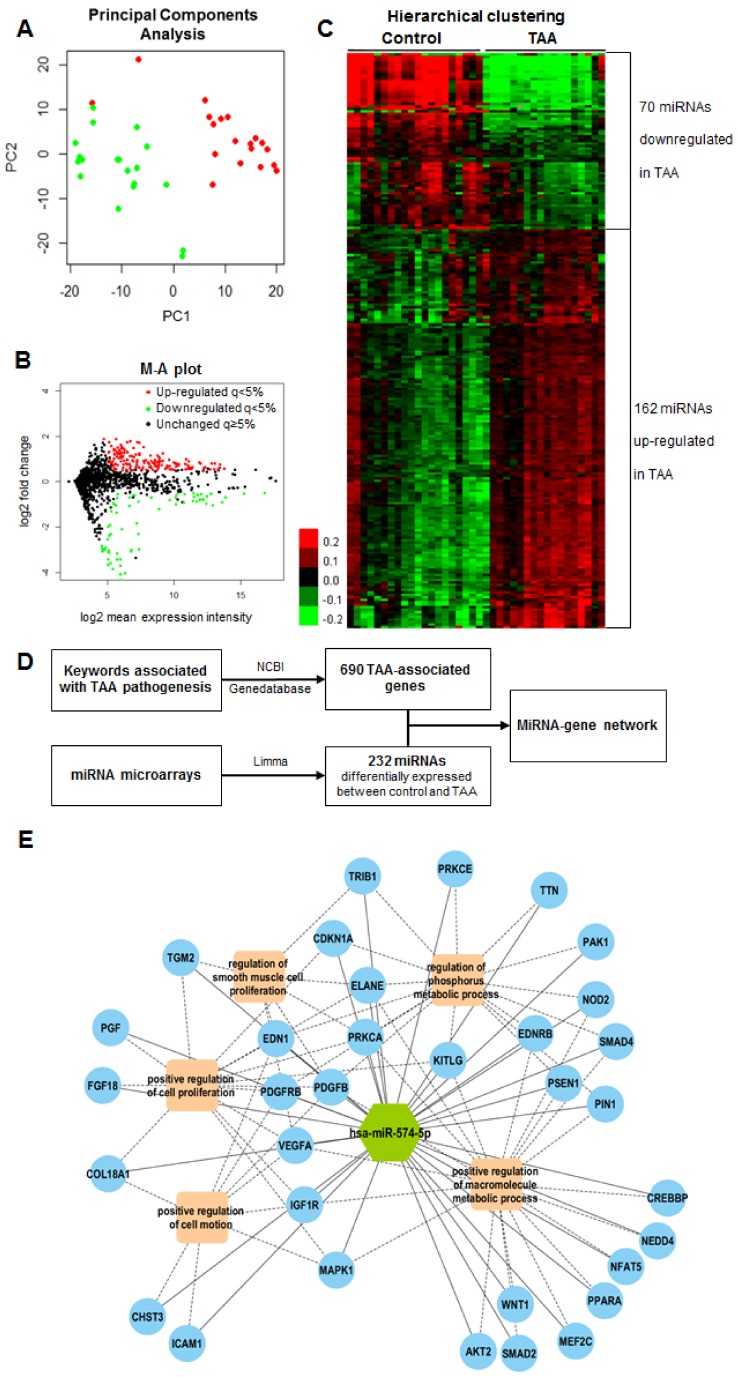
Figure 1.
MicroRNAs (MiRNA) expression profiles in aortic tissues from the tissue cohort and miRNA-gene and protein-protein interaction network. (A–C). Microarrays were conducted on RNA samples isolated from the aortic media of 19 thoracic aortic aneurysm (TAA) patients and 19 non-aneurysmal control donors. (A) The principal component analysis showing the separation of TAA and control samples according to miRNA expression. (B) M-A plot showing the distribution of miRNAs in TAA versus control samples. Log2 transformed-fold change is shown in the vertical axis and the average signal of each miRNA is shown in the horizontal axis. (C) Heat-map displaying modulated miRNAs between TAA and control samples. A log2-transformed fold change > 0.5 or < −0.5, a q-value <5% and a relative expression level > 5 were used as cut-offs. Red color corresponds to miRNAs up-regulated in TAA patients and green color corresponds to miRNAs down-regulated in TAA patients compared to controls. (D,E) MiRNA-gene and protein-protein interaction network. (D) Procedure followed to create the interaction network. 690 TAA-associated genes were identified using the National Center for Biotechnology Information (NCBI) Gene database and literature search. 232 miRNAs were found to be differentially expressed between aneurysmal and non-aneurysmal aortic tissue using microarrays. Interactions between TAA-associated genes and differentially expressed miRNAs were predicted using TargetScan, microRNA Target Prediction Database (miRDB) and DIANA microT-CDS databases. Only the interactions predicted by at least two databases were considered. Functional protein-protein interactions identified in the IntAct database were also included in the network. (E) MiR-574-5p-gene interaction network. The network shows the association between-miR-574-5p, its predicted gene targets, and the top five biological processes enriched by these targets. Solid line: Interaction between miRNA and the target gene. Dash line: Association between gene targets and biological processes.