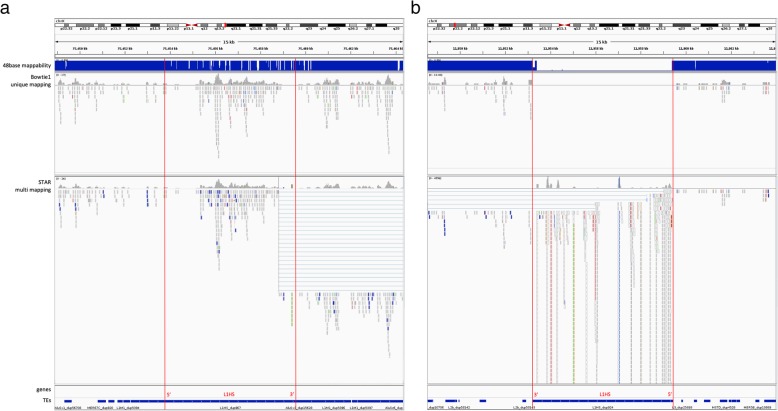
Fig. 1.
Comparison of read alignment on full length L1HS with multi-mapping and unique mapping. Reads mapped to two different full length L1HS elements from a stomach tissue sample (A4GY) visualized through IGV. a L1HS_dup967, a 5799 nt length element on chromosome X: 75453754–75459553. b a 6225 nt length element L1HS_dup924 on chromosome X: 11953208–11959433. Chromosomal locations, 48 base mappability calculated with GEM, Bowtie1 alignment allowing only uniquely mapped reads with single best hit, STAR alignment allowing multi-mapped reads up to 200 mapping for each read, gene annotation and Repeatmasker TE annotation are shown from top to bottom. Red lines mark the boundary of the L1HS elements with 5′ and 3′ noted