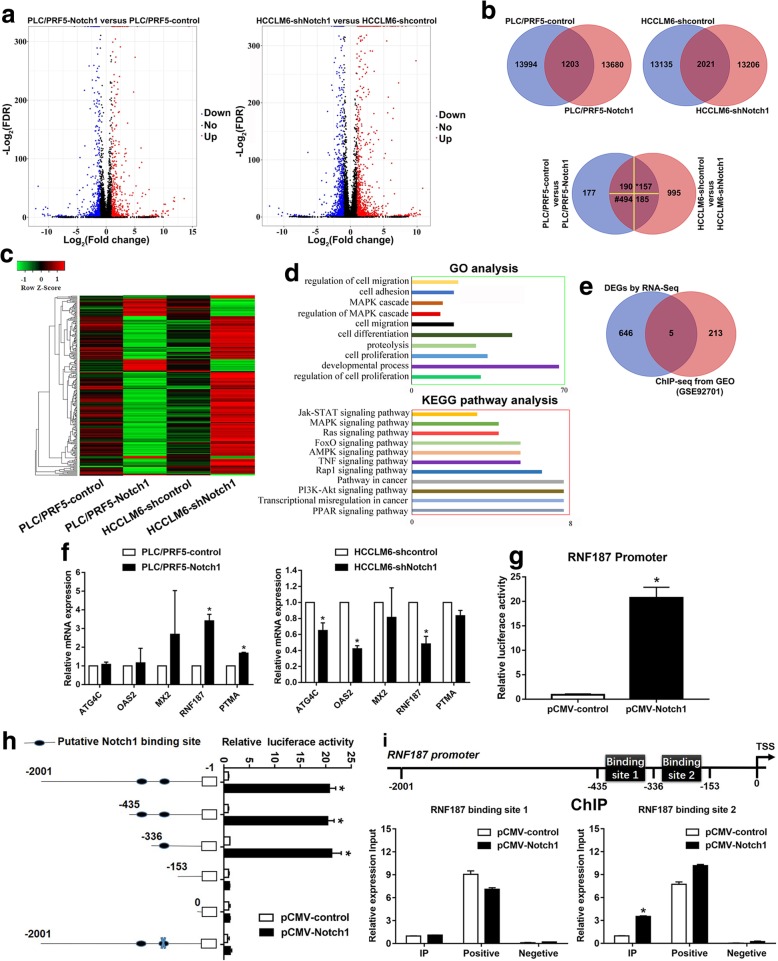
Fig. 4.
RNF187 is a downstream target of Notch1. a The scatter plot showed significant differentially expression genes (DEGs) of Notch1-modulated cells by RNA-seq; (b) Venn analysis was used to screen overlapping mRNA in step a; *: up-regulation in PLC/PRF5-Notch1 versus control group, simultaneously, down-regulation in HCCLM6-shNotch1 versus shcontrol group; #: down-regulation in PLC/PRF5-Notch1 versus control group, simultaneously, up-regulation in HCCLM6-shNotch1 versus shcontrol group. c Heat map and (d) GO categories, KEGG pathways enriched in the 651 DEGs from step b. e Venn diagrams to show intersections between RNA-seq gene sets (651 DEGs from step b) and ChIP-sequencing of Notch1 database (GEO accession no. GSE92701). f Real-time PCR was used to verify the DEGs. g Notch1 transactivates RNF187 promoter activity, as detected using a luciferase reporter assay. h Deletion and selective mutation analysis identified Notch responsive region in the RNF187 promoter, and relative luciferase activity was measured. i ChIP assay identified Notch1-binding sites in the RNF187 promoter. Real-time PCR was performed to detect the amounts of immunoprecipitated products. The positive control: anti-RNA polymerase II; the negative control: normal mouse IgG. *: P < 0.05