Figure 1.
Temporal Proteomic Profiling of EBV B Cell Transformation Reveals Dramatic Metabolic Remodeling
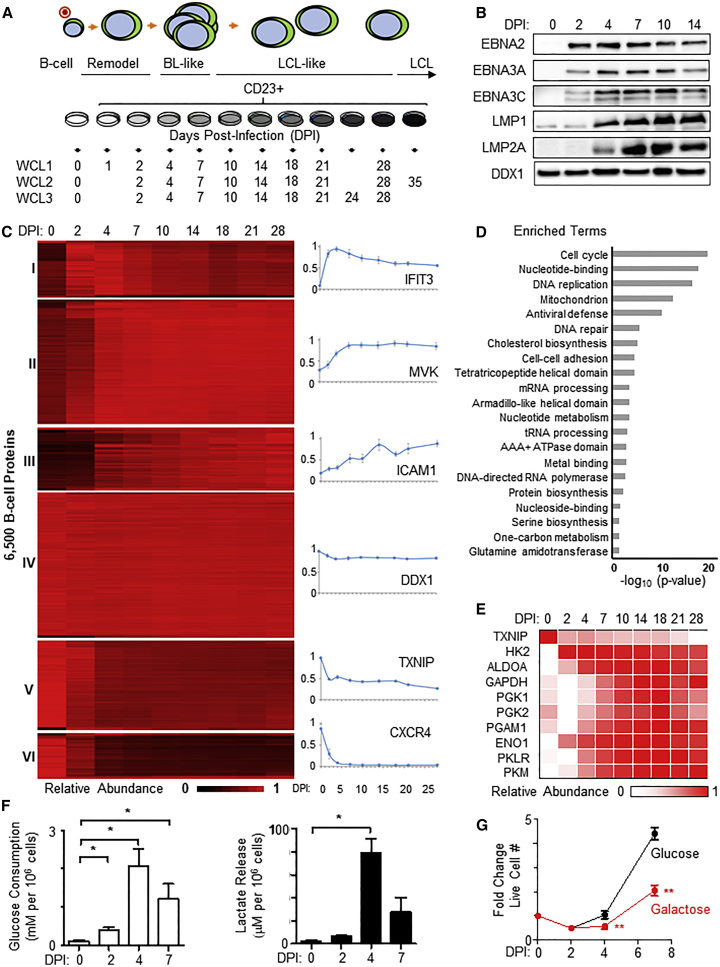
(A) Schematic of experimental workflow. Primary human B cells (3 biological replicates, each consisting of 4 independent donors) were analyzed prior to and at nine time points after EBV infection. CD23+ EBV-infected cells were enriched by fluorescence-activated cell sorting (FACS). WCL, whole-cell lysate.
(B) Representative immunoblots (n = 3) of EBV nuclear antigens (EBNAs), latent membrane proteins (LMPs), and load-control DDX1 of newly infected primary B cells at the indicated days post-infection (DPI).
(C) k-means clustering (k = 6) of the ∼6,500 B cell proteins quantified in all three replicates based on averaged relative abundances. Representative expression profiles are shown to the right of each k-means cluster. To determine the actual number of distinct classes of host protein expression, the k-means approach was used with 1–12 classes to cluster viral proteins, and the summed distance of each protein from its cluster centroid was calculated (see Figure S1D).
(D) Functional enrichment within all proteins upregulated >2-fold with p < 0.075 4 days post-EBV infection (clusters I–III) against a background of all quantified proteins. Overall, 41 clusters were significantly enriched (Table S2). For the purposes of simplified display, these were concatenated into hierarchical parent terms, where available for individual clusters that had been identified using Uniprot or Gene Ontology. For example, “Sister chromatid cohesion,” “DNA condensation,” and “Kinesin, motor domain” were concatenated into “Cell cycle.”
(E) Heatmap of averaged relative abundances of glycolytic pathway component enzymes at each indicated time point.
(F) LC-MS analysis of media glucose consumption and lactate production from cultures of primary B cells at the indicated time points post-infection. The 24-h mean ± SEM decrease in media glucose and 24-h increase in media lactate are shown at the indicated DPI, n = 3. ∗p < 0.05 (paired one-tailed t test).
(G) Growth curves of newly infected primary human B cells grown in complete media containing either glucose or galactose. Data show the mean ± SEM, n = 3. ∗∗p < 0.01 (paired one-tailed t test).
See also Figures S1–S3 and Tables S1 and S2.