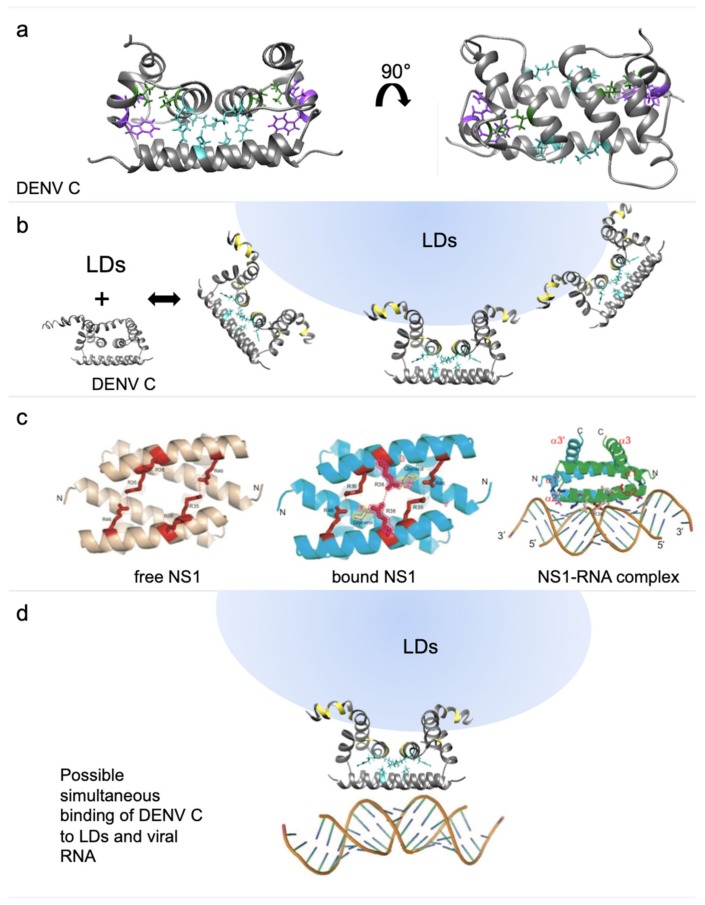
Figure 7.
Protein structures of DENV C and influenza NS1. (a) DENV C structure from two different angles with the conserved residues R68 and W69 (purple) and the interface stabilizing residues V51 and I65 (green), as well as E87, R55 and K45, forming the salt bridge (cyan). (b) DENV C structure in a N-terminal region closed conformation and, next, in an open conformation with schematic binding of lipid droplets (LDs) and the affected amino acid residues (yellow). (c) The RNA-binding domain of NS1 protein from influenza A in a RNA-free (left) and RNA-bound state (middle and right), showing an organization similar to DENV C α4–α4′ region (adapted from Cheng et al., 2009 [44]). (d) DENV C with schematically bound to a LD and to RNA. DENV C amino acid residues affected by the binding to LDs are colored yellow, while a key internal salt bridge is shown in cyan. DENV C binding to host LDs may enable allosteric rearrangements (eventually involving the salt bridge), allowing a small conformational change in α4 side chains, namely the positively charged residues, prompting stable RNA-C protein binding.