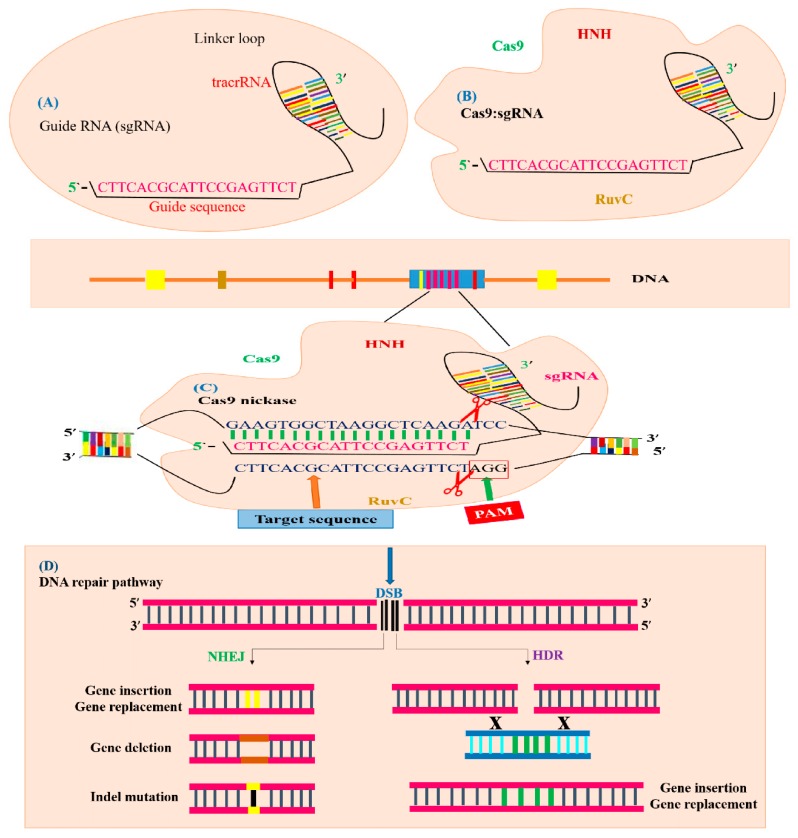
Figure 5.
Illustration of CRISPR/Cas9-mediated GE. The CRISPR/Cas9 system is composed of sgRNA and Cas9. (A) sgRNA with a guide sequence (colored pink) is developed by the combination of protospacer with crRNA and tracrRNA. (B) Cas9 machinery combines with sgRNA to form a complex to trigger CRISPR/Cas9 editing. The Cas9 nuclease consists of two parts, depending on its function and structure. The recognition site identifies the target DNA and interacts with sgRNA. The nuclease site contains two domains RuvC-like and HNH which cleave the target DNA site non-complementary by the RuvC domain and complementary by the HNH domain to the gRNA. (C) The Cas9 nuclease detects the genomic target site (indicated with blue color) having a 20 bp target sequence that is homologous to seed or guide sequence (indicated with pink color), which is crucial for Cas9 activity and specificity. The specific PAM sequence (indicated with red color) is detected by Cas9: sgRNA complex and DSBs created by the Cas9 endonuclease three base pairs upstream of the PAM sequence. (D) Targeted mutagenesis of a desired gene is achieved by filling the DSB (indicated with black color) by means of the HDR or NHEJ mechanism. The NHEJ repair mechanism generally produces insertion (indicated with yellow color), deletion (indicated with brown color) or indels (indicated with black line) at the break point, generating targeted mutants. The HDR repair mechanism uses a template DNA sequence for homologous recombination to produce gene replacement or gene insertion (indicated with green color).